| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,276,764 – 21,276,828 |
| Length | 64 |
| Max. P | 0.910208 |

| Location | 21,276,764 – 21,276,828 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Shannon entropy | 0.39946 |
| G+C content | 0.34222 |
| Mean single sequence MFE | -10.61 |
| Consensus MFE | -6.21 |
| Energy contribution | -7.13 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910208 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
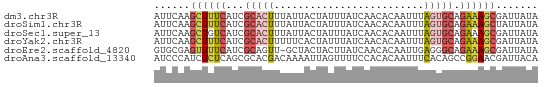
>dm3.chr3R 21276764 64 + 27905053 AUUCAAGCUUUCAUCGCACUUUAUUACUAUUUAUCAACACAAUUUAGUGCAGAAAGCGAUUAUA ......((((((...(((((..(((...............)))..))))).))))))....... ( -12.96, z-score = -3.41, R) >droSim1.chr3R 21104938 64 + 27517382 AUUCAAGCUUUCAUCGCACUUUAUUACUAUUUAUCAACACAAUUUAGUGCAGAAAGCUAUUAUA .....(((((((...(((((..(((...............)))..))))).)))))))...... ( -13.66, z-score = -4.16, R) >droSec1.super_13 62635 64 + 2104621 AUUCAAGCUGUCAUCGCACUUUAUUACUAUUUAUCAACACAAUUUAGUGCAGAAAGCGAUUAUA ......(((.((...(((((..(((...............)))..))))).)).)))....... ( -9.46, z-score = -1.35, R) >droYak2.chr3R 3623873 64 - 28832112 AUUCAAGCUUUCAUCGCACUUUUUCACUAUUUAUCAACACAAUUUAGUGCAGAAAGCGAUUAUA ......((((((...(((((.........................))))).))))))....... ( -12.01, z-score = -3.72, R) >droEre2.scaffold_4820 3701714 63 - 10470090 GUGCGAGUUUUCAUCGCAGUU-GCUACUACUUAUCAACACAAUUGAGGGCAGAAAGCGAUUAUA ..((((.......))))((((-(((.((.(((.((((.....))))))).))..)))))))... ( -12.90, z-score = -0.72, R) >droAna3.scaffold_13340 14494146 64 - 23697760 AUCCCAUCUCUCAGCGCACGACAAAAUUAGUUUUCCACACAAUUUCACAGCCGGAACGAUUACA ...........................(((((((((................)))).))))).. ( -2.69, z-score = 1.00, R) >consensus AUUCAAGCUUUCAUCGCACUUUAUUACUAUUUAUCAACACAAUUUAGUGCAGAAAGCGAUUAUA ......((((((...(((((.........................))))).))))))....... ( -6.21 = -7.13 + 0.92)

| Location | 21,276,764 – 21,276,828 |
|---|---|
| Length | 64 |
| Sequences | 6 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Shannon entropy | 0.39946 |
| G+C content | 0.34222 |
| Mean single sequence MFE | -13.93 |
| Consensus MFE | -7.30 |
| Energy contribution | -7.75 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.770768 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 21276764 64 - 27905053 UAUAAUCGCUUUCUGCACUAAAUUGUGUUGAUAAAUAGUAAUAAAGUGCGAUGAAAGCUUGAAU .......((((((((((((..((((((((....)))).))))..))))))..))))))...... ( -16.00, z-score = -3.09, R) >droSim1.chr3R 21104938 64 - 27517382 UAUAAUAGCUUUCUGCACUAAAUUGUGUUGAUAAAUAGUAAUAAAGUGCGAUGAAAGCUUGAAU ......(((((((((((((..((((((((....)))).))))..))))))..)))))))..... ( -16.40, z-score = -3.37, R) >droSec1.super_13 62635 64 - 2104621 UAUAAUCGCUUUCUGCACUAAAUUGUGUUGAUAAAUAGUAAUAAAGUGCGAUGACAGCUUGAAU .......(((.((((((((..((((((((....)))).))))..))))))..)).)))...... ( -12.50, z-score = -1.42, R) >droYak2.chr3R 3623873 64 + 28832112 UAUAAUCGCUUUCUGCACUAAAUUGUGUUGAUAAAUAGUGAAAAAGUGCGAUGAAAGCUUGAAU .......((((((((((((..(((((........))))).....))))))..))))))...... ( -15.30, z-score = -3.20, R) >droEre2.scaffold_4820 3701714 63 + 10470090 UAUAAUCGCUUUCUGCCCUCAAUUGUGUUGAUAAGUAGUAGC-AACUGCGAUGAAAACUCGCAC .......(((..((((..(((((...)))))...)))).)))-...((((((....).))))). ( -13.70, z-score = -1.66, R) >droAna3.scaffold_13340 14494146 64 + 23697760 UGUAAUCGUUCCGGCUGUGAAAUUGUGUGGAAAACUAAUUUUGUCGUGCGCUGAGAGAUGGGAU .....((((((((((.(..((((((.((.....))))))))..).....)))).)).))))... ( -9.70, z-score = 0.76, R) >consensus UAUAAUCGCUUUCUGCACUAAAUUGUGUUGAUAAAUAGUAAUAAAGUGCGAUGAAAGCUUGAAU .......((((((((((((..(((((........))))).....))))))..))))))...... ( -7.30 = -7.75 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:14 2011