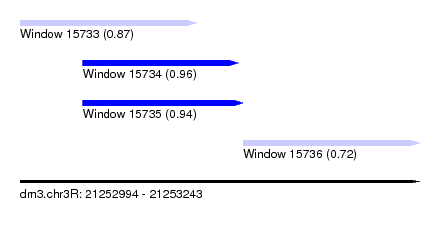
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,252,994 – 21,253,243 |
| Length | 249 |
| Max. P | 0.955508 |

| Location | 21,252,994 – 21,253,104 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.04 |
| Shannon entropy | 0.11165 |
| G+C content | 0.42292 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21252994 110 + 27905053 AAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCA ...........(((...-....(((((..((..((....)).))..)))))........(-((((((((((....))))))))))).....))).................. ( -25.60, z-score = -2.81, R) >droAna3.scaffold_13340 14474430 112 - 23697760 AAUUGGUUUCCGCCUCGCUUUUUAUAACCCUUUCAACUUUGUGGAUUUAUGUUCCAUUUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGGCAAUAAAAUAUAUAUGCGA ....(((....)))((((....((((..............(((((.......)))))...((((((....))))))(((..((.....)).))).......))))...)))) ( -25.10, z-score = -0.29, R) >droEre2.scaffold_4820 3678439 110 - 10470090 AAUUACUUUCCGGCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCA ...........((...(-((.....)))...........((((.((((.(((((((...(-((((((((((....)))))))))))....))).)))))))).))))..)). ( -26.30, z-score = -2.79, R) >droYak2.chr3R 3599838 110 - 28832112 AAUUACUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCCGUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUAGCA ...........((...(-((.....)))............(((.((((.(((((((...(-((((((((((....)))))))))))....))).)))))))).)))...)). ( -25.80, z-score = -2.70, R) >droSec1.super_13 26869 110 + 2104621 AAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUACAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCA ...........(((..(-(...(((((..((..((....)).))..)))))..))....(-((((((((((....))))))))))).....))).................. ( -27.20, z-score = -3.23, R) >droSim1.chr3R 21081429 110 + 27517382 AAUUGCUUUCCGCCUCG-UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCCGGCAAUAAAAUAUAUAUACCA ...........(((...-....(((((..((..((....)).))..)))))........(-((((((((((....))))))))))).....))).................. ( -25.90, z-score = -2.86, R) >consensus AAUUGCUUUCCGCCUCG_UUUUUAUAACCCUUUCAACUCUGUAGAUUUAUGUUCCAUUUU_AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCA ...........(((........(((((..((..((....)).))..)))))..........((((((((((....))))))))))......))).................. (-20.50 = -21.03 + 0.53)
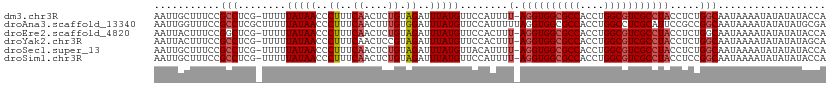
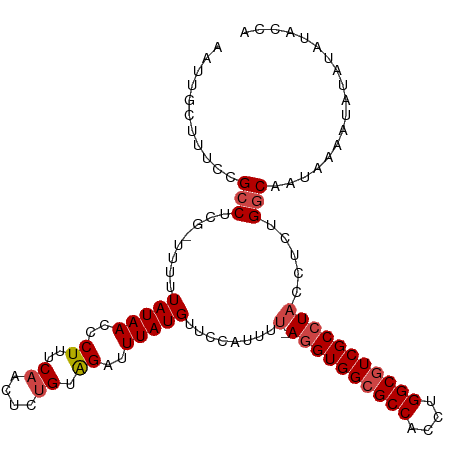
| Location | 21,253,033 – 21,253,130 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.49 |
| Shannon entropy | 0.19662 |
| G+C content | 0.41936 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.52 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
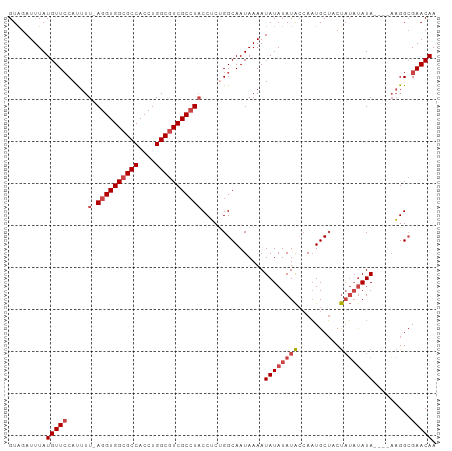
>dm3.chr3R 21253033 97 + 27905053 GUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAA .........(((((.....(-((((((((((....)))))))))))......((.......((((((((..........))))))))----...))))))). ( -28.00, z-score = -2.65, R) >droAna3.scaffold_13340 14474470 100 - 23697760 GUGGAUUUAUGUUCCAUUUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGGCAAUAAAAUAUAUAUGCGAUUGCACCCCGCCAUAUC--CCGGAGAACAA .((((.......))))((((..(((((((((....))).((((((.....................)))))).........))))...)--)..)))).... ( -22.90, z-score = 0.95, R) >droEre2.scaffold_4820 3678478 99 - 10470090 GUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUUCUAUAUAUAUA--AAGGCGAACAA .........(((((.....(-((((((((((....)))))))))))......((......(((((((((..........))))))))).--...))))))). ( -30.10, z-score = -3.42, R) >droYak2.chr3R 3599877 101 - 28832112 GUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUAGCAAUGCUCCUAUAUAUAUAUAAAGGCGAACAA .........(((((.....(-((((((((((....)))))))))))......((......((((((((((........))))))))))......))))))). ( -34.40, z-score = -4.15, R) >droSec1.super_13 26908 97 + 2104621 GUAGAUUUAUGUUACAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAA ((...(((((((.......(-((((((((((....)))))))))))....(((((..................)))))....)))))----)).))...... ( -26.07, z-score = -2.01, R) >droSim1.chr3R 21081468 97 + 27517382 GUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCCGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAA .........(((((.....(-((((((((((....)))))))))))......((.......((((((((..........))))))))----...))))))). ( -28.00, z-score = -2.60, R) >consensus GUAGAUUUAUGUUCCAUUUU_AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA____AAGGCGAACAA .........(((((.......((((((((((....)))))))))).......((.......((((((((..........)))))))).......))))))). (-23.47 = -24.52 + 1.06)

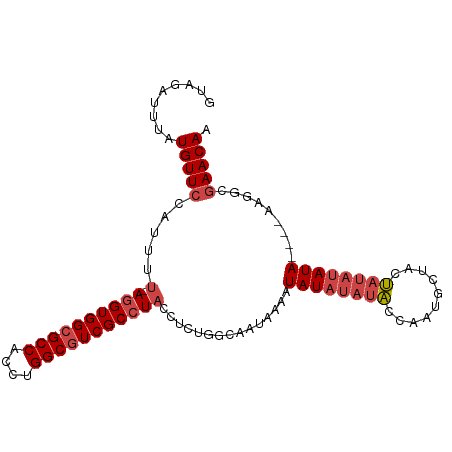
| Location | 21,253,033 – 21,253,133 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.47 |
| Shannon entropy | 0.19719 |
| G+C content | 0.40859 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.52 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
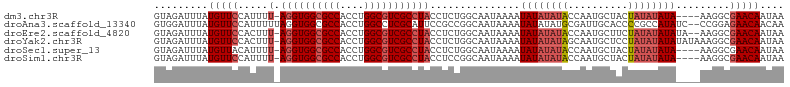
>dm3.chr3R 21253033 100 + 27905053 GUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAAUAA .........(((((.....(-((((((((((....)))))))))))......((.......((((((((..........))))))))----...))))))).... ( -28.00, z-score = -2.55, R) >droAna3.scaffold_13340 14474470 103 - 23697760 GUGGAUUUAUGUUCCAUUUUUAGGUGGCGCCACCUGGCCUCGCAUUCCGCCGGCAAUAAAAUAUAUAUGCGAUUGCACCCCGCCAUAUC--CCGGAGAACAACAA .((((.......))))((((..(((((((((....))).((((((.....................)))))).........))))...)--)..))))....... ( -22.90, z-score = 0.95, R) >droEre2.scaffold_4820 3678478 102 - 10470090 GUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUUCUAUAUAUAUA--AAGGCGAACAAUAA .........(((((.....(-((((((((((....)))))))))))......((......(((((((((..........))))))))).--...))))))).... ( -30.10, z-score = -3.29, R) >droYak2.chr3R 3599877 104 - 28832112 GUAGAUUUAUGUUCCACUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUAGCAAUGCUCCUAUAUAUAUAUAAAGGCGAACAAUAA .........(((((.....(-((((((((((....)))))))))))......((......((((((((((........))))))))))......))))))).... ( -34.40, z-score = -4.04, R) >droSec1.super_13 26908 100 + 2104621 GUAGAUUUAUGUUACAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAAUAA ((...(((((((.......(-((((((((((....)))))))))))....(((((..................)))))....)))))----)).))......... ( -26.07, z-score = -1.94, R) >droSim1.chr3R 21081468 100 + 27517382 GUAGAUUUAUGUUCCAUUUU-AGGUGGCGCCACCUGGCGUCGCCUACCUCCGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA----AAGGCGAACAAUAA .........(((((.....(-((((((((((....)))))))))))......((.......((((((((..........))))))))----...))))))).... ( -28.00, z-score = -2.52, R) >consensus GUAGAUUUAUGUUCCAUUUU_AGGUGGCGCCACCUGGCGUCGCCUACCUCUGGCAAUAAAAUAUAUAUACCAAUGCUACUAUAUAUA____AAGGCGAACAAUAA .........(((((.......((((((((((....)))))))))).......((.......((((((((..........)))))))).......))))))).... (-23.47 = -24.52 + 1.06)
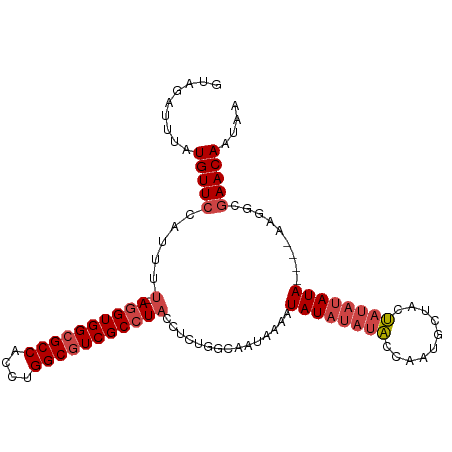
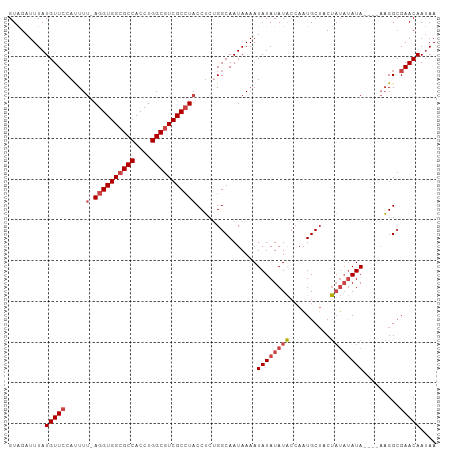
| Location | 21,253,133 – 21,253,243 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Shannon entropy | 0.24880 |
| G+C content | 0.41187 |
| Mean single sequence MFE | -18.59 |
| Consensus MFE | -15.33 |
| Energy contribution | -15.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21253133 110 + 27905053 AAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCAGCUGACAGCAAAAAA-UG ......((((((......................((.......)).(((((((........))))))).))))))..................((.....))......-.. ( -18.60, z-score = -1.43, R) >droSim1.chr3R 21081568 110 + 27517382 AAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAGCAAAAAA-UG ......((((((......................((.......)).(((((((........))))))).)))))).................................-.. ( -17.20, z-score = -1.29, R) >droSec1.super_13 27008 110 + 2104621 AAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAG-AAAAAAAUG ......((((((......................((.......)).(((((((........))))))).))))))..........................-......... ( -17.20, z-score = -1.56, R) >droYak2.chr3R 3599981 111 - 28832112 AAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUUACUCAGCCGAGAAGUUAAAUACAAUCUGCCACCCACUGACAACAAAAAAAUG .................................(((((........(((((((........)))))))....((.....))...)))))...................... ( -18.20, z-score = -1.89, R) >droEre2.scaffold_4820 3678580 111 - 10470090 AAACACAACUUCAUACACGACAUAAAUCAAGCGGGCAGUUCAAGCAUGGCUGAAAUAUUACUCAGCCGAGAAGUUAAAUACAAACUGCCACCCACUGACAACAAAAAAAUG ..........................(((.(..(((((((......(((((((........)))))))....((.....)).)))))))..)...)))............. ( -20.80, z-score = -2.56, R) >droAna3.scaffold_13340 14474573 97 - 23697760 AAGCAAUAUUCCAUGCACGACAUAAAUCACCUGGGCAGUUCAAGCCGGGCUGAAUUAUUACUCGGCCGAGAAGUUAAAUAAAAGCCCGCAAAAAAUG-------------- .............(((.................(((.......)))(((((.....(((.(((....))).)))........)))))))).......-------------- ( -19.52, z-score = -0.26, R) >consensus AAACACAACUUCAUACACGACAUAAAUCAACCGGGCAGUUCAAGCAUGGCUGAAAUAUAACUCAGCCGAGAAGUUAAAUACAAACCGCCACCCACUGACAGCAAAAAA_UG ......((((((......................((.......))..((((((........))))))..)))))).................................... (-15.33 = -15.38 + 0.06)

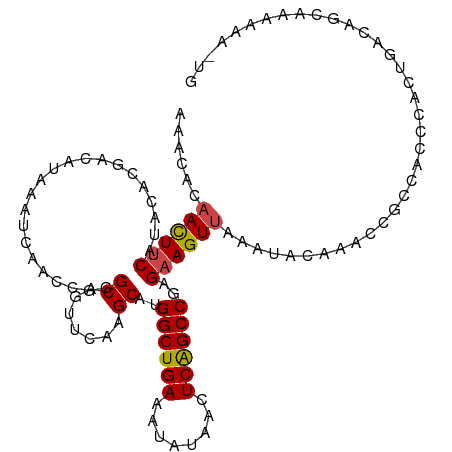
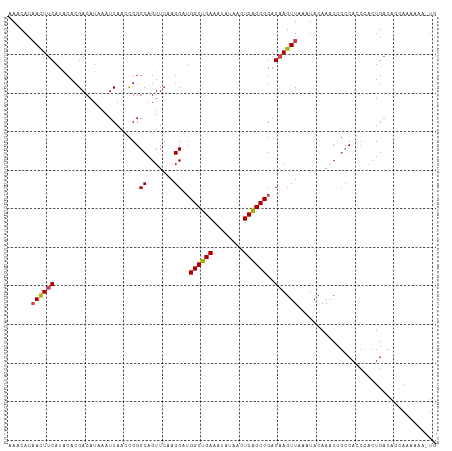
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:10 2011