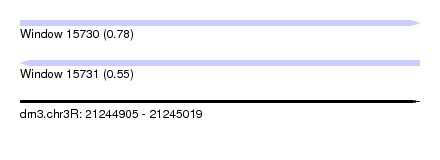
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,244,905 – 21,245,019 |
| Length | 114 |
| Max. P | 0.779751 |

| Location | 21,244,905 – 21,245,019 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.94 |
| Shannon entropy | 0.40194 |
| G+C content | 0.45687 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -17.29 |
| Energy contribution | -19.68 |
| Covariance contribution | 2.39 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
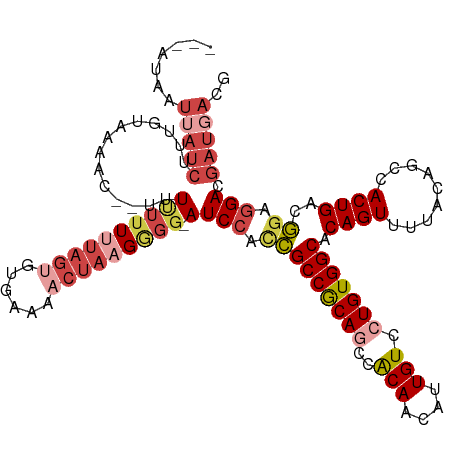
>dm3.chr3R 21244905 114 + 27905053 ---AUAAUUAUCUUUGAAAAUA--UAUUUUUUUAGUGUGAAAACUAAGGGU-AUCCCCCGCCGCAGCCACAACAUUGUCCUGUGGCACAGUUUUACAGCCACUGACGGACGACGAUGACG ---....(((((.(((((((..--...))))((((((((((((((..(((.-....)))(((((((..(((....))).)))))))..)))))).))..))))))....))).))))).. ( -28.50, z-score = -2.04, R) >droAna3.scaffold_13340 14467482 105 - 23697760 -------------UUGAUGGCCA-AUUCGGUUUAAAGUGUAGCCGAAGAGGCAUCCAUGGCCACAGCCGCAACAUUGUGCUGUGGCACAGUUUUGCAGCCACUGAAUG-GGACGUUGUCG -------------..(((((((.-.(((((((........)))))))..))).(((((.((((((((.(((....))))))))))).((((.........)))).)))-)).)))).... ( -40.00, z-score = -2.17, R) >droEre2.scaffold_4820 3670086 114 - 10470090 AUUUUUAUAAUCUUUGUAAG----UCUUUUUUUAGUGUGAAAUCUA-GGGG-AUCCAACGCCGCAUCCACAACAUUGUACUGUGGCACAGUUUUACAGCCACUGACGGAGGACGAUGACG ...............((..(----(((((..((((((((((((...-((..-..))...((((((...(((....)))..))))))...)))))))....)))))..))))))....)). ( -26.60, z-score = -0.70, R) >droYak2.chr3R 3591298 111 - 28832112 ------AUUAUCUUUGUAGACC--UUUUUUUAAAGUGUCCAAACUAGGAGG-AUCCACCGCCGCAUCCACAACAUUGUCCUGUGGCACAGUUUUACAGCCACUGACGGAGGACGAUGACG ------....((((.((.((((--(((....)))).)))...)).))))((-((.(......).))))....((((((((((((((...........)))))......)))))))))... ( -28.80, z-score = -1.57, R) >droSec1.super_13 18466 119 + 2104621 AUUAUAAUUAUCUUUGUGCAACCUUUUUUUUUUAGUGUGAACACUAAGGGG-AUCCCCCGCCGCAGCCACAACAUUGUCCUGUGGCACAGUUUUACAGCCACUGACGGAGGACGAUGACG .............((((((............((((((....))))))((((-...))))......).)))))((((((((((((((...........)))))......)))))))))... ( -35.90, z-score = -2.95, R) >droSim1.chr3R 21072655 117 + 27517382 AUUAUAAUUAUCUUUGUAUAAC--AUUUUUUUUAGUGUGAAAACUAAGGGG-AUCCCCCGCCGCAGCCACAACAUUGUCCUGUGGCACAGUUUUACAGCCACUGACGGAGGACGAUGACG .......(((((..((.....)--)((((..((((((((((((((..((((-...))))(((((((..(((....))).)))))))..)))))).))..))))))..))))..))))).. ( -31.60, z-score = -2.06, R) >consensus ___AUAAUUAUCUUUGUAAAAC__UUUUUUUUUAGUGUGAAAACUAAGGGG_AUCCACCGCCGCAGCCACAACAUUGUCCUGUGGCACAGUUUUACAGCCACUGACGGAGGACGAUGACG .......(((((................((((((((......))))))))...(((.(((((((((..(((....))).))))))).((((.........))))..)).))).))))).. (-17.29 = -19.68 + 2.39)

| Location | 21,244,905 – 21,245,019 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.94 |
| Shannon entropy | 0.40194 |
| G+C content | 0.45687 |
| Mean single sequence MFE | -31.79 |
| Consensus MFE | -18.43 |
| Energy contribution | -19.32 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
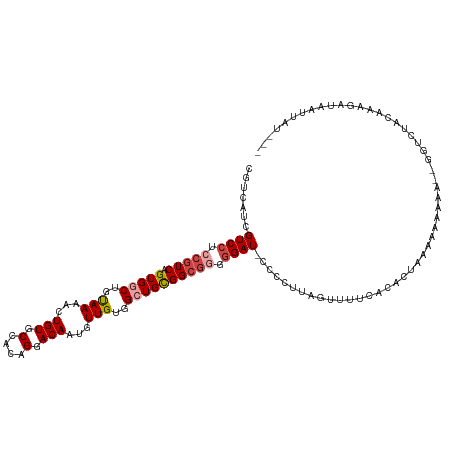
>dm3.chr3R 21244905 114 - 27905053 CGUCAUCGUCGUCCGUCAGUGGCUGUAAAACUGUGCCACAGGACAAUGUUGUGGCUGCGGCGGGGGAU-ACCCUUAGUUUUCACACUAAAAAAAUA--UAUUUUCAAAGAUAAUUAU--- .(((...(((.((((((.(..((..(((...(((.(....).)))...)))..))..))))))).)))-....(((((......))))).......--..........)))......--- ( -29.50, z-score = -1.15, R) >droAna3.scaffold_13340 14467482 105 + 23697760 CGACAACGUCC-CAUUCAGUGGCUGCAAAACUGUGCCACAGCACAAUGUUGCGGCUGUGGCCAUGGAUGCCUCUUCGGCUACACUUUAAACCGAAU-UGGCCAUCAA------------- .......(.((-.((((.((((((((((...(((((....)))))...))))))))((((((..(((....)))..)))))).......)).))))-.)).).....------------- ( -36.00, z-score = -2.09, R) >droEre2.scaffold_4820 3670086 114 + 10470090 CGUCAUCGUCCUCCGUCAGUGGCUGUAAAACUGUGCCACAGUACAAUGUUGUGGAUGCGGCGUUGGAU-CCCC-UAGAUUUCACACUAAAAAAAGA----CUUACAAAGAUUAUAAAAAU .((.(((.......(((.(((((...........)))))(((.(((((((((....)))))))))(((-(...-..))))....))).......))----).......))).))...... ( -22.64, z-score = -0.02, R) >droYak2.chr3R 3591298 111 + 28832112 CGUCAUCGUCCUCCGUCAGUGGCUGUAAAACUGUGCCACAGGACAAUGUUGUGGAUGCGGCGGUGGAU-CCUCCUAGUUUGGACACUUUAAAAAAA--GGUCUACAAAGAUAAU------ .(((...((((.(((((.(((((...........)))))..)))...(((((....))))))).))))-..........(((((.((((....)))--))))))....)))...------ ( -30.90, z-score = -1.04, R) >droSec1.super_13 18466 119 - 2104621 CGUCAUCGUCCUCCGUCAGUGGCUGUAAAACUGUGCCACAGGACAAUGUUGUGGCUGCGGCGGGGGAU-CCCCUUAGUGUUCACACUAAAAAAAAAAGGUUGCACAAAGAUAAUUAUAAU .((.(((((((((((((.(..((..(((...(((.(....).)))...)))..))..)))))))))))-....((((((....))))))........))).))................. ( -38.40, z-score = -2.70, R) >droSim1.chr3R 21072655 117 - 27517382 CGUCAUCGUCCUCCGUCAGUGGCUGUAAAACUGUGCCACAGGACAAUGUUGUGGCUGCGGCGGGGGAU-CCCCUUAGUUUUCACACUAAAAAAAAU--GUUAUACAAAGAUAAUUAUAAU .(((...((((((((((.(..((..(((...(((.(....).)))...)))..))..)))))))))))-....(((((......))))).......--..........)))......... ( -33.30, z-score = -1.94, R) >consensus CGUCAUCGUCCUCCGUCAGUGGCUGUAAAACUGUGCCACAGGACAAUGUUGUGGCUGCGGCGGGGGAU_CCCCUUAGUUUUCACACUAAAAAAAAA__GGUCUACAAAGAUAAUUAU___ .......((((.(((((.(((((..(((...(((.(....).)))...)))..)))))))))).)))).................................................... (-18.43 = -19.32 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:06 2011