
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,237,973 – 21,238,071 |
| Length | 98 |
| Max. P | 0.829656 |

| Location | 21,237,973 – 21,238,071 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 65.40 |
| Shannon entropy | 0.71112 |
| G+C content | 0.46687 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -7.39 |
| Energy contribution | -7.27 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21237973 98 + 27905053 CCCAGCAAACACAGGCACACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCUCGGGC---ACGCCCACAACAUCAAAGGGCGUCAUACU-GACA-UUGA--- ....((........)).......................(((--((........(((...)))---((((((...........))))))...)))-))..-....--- ( -17.70, z-score = -1.31, R) >droSim1.chr3R 21065580 98 + 27517382 CCCAGCAAACACAGGCACACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCUCGGGC---ACGCCCACAACAUCAAAGGGCGUCAUACU-GACA-UUGA--- ....((........)).......................(((--((........(((...)))---((((((...........))))))...)))-))..-....--- ( -17.70, z-score = -1.31, R) >droSec1.super_13 11526 98 + 2104621 CGCAGCAAACGCAGGCACACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCUCCGGC---ACGCCCACAACAUCAAAGGGCGUCAUACU-GACA-UUGA--- .((.((....))..)).......................(((--((........(((...)))---((((((...........))))))...)))-))..-....--- ( -19.70, z-score = -1.56, R) >droYak2.chr3R 3584153 98 - 28832112 CCCAGCAAACACAGGCACACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCCCGGGC---ACGCCCACAACAUCAAAGGGCGUCAUACU-GACA-UUGA--- ....((........)).......................(((--((........(((...)))---((((((...........))))))...)))-))..-....--- ( -17.70, z-score = -1.09, R) >droEre2.scaffold_4820 3662959 98 - 10470090 GCCAGCAAACACAGGCACACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCUCGGGC---ACGCCCACAACAUCAAAGGGCGUCAUACU-GACA-UUGA--- (((..........))).......................(((--((........(((...)))---((((((...........))))))...)))-))..-....--- ( -20.00, z-score = -2.11, R) >droAna3.scaffold_13340 14460348 94 - 23697760 ----CACAGCGCACAACCACAUUCAAUUACACAUAAGAUUCA--GUUUACUUUCGCCUCGGGC---CCGCCCACGACAUCAAAGGGCGUCGCUCU-GACA-UUGA--- ----..(((.((..............................--..........(((...)))---.(((((...........)))))..)).))-)...-....--- ( -16.90, z-score = -0.15, R) >dp4.chr2 3324772 92 - 30794189 ----------ACACGCCCACAUUCAAUUACAGUGAAGAUUUGCCGUUUACUUUUCCCCCAGGCC--ACGCCCACAACAUCAAAGGGCGUUGACUUAGACA-UUGA--- ----------...................(((((.......(((................))).--((((((...........)))))).........))-))).--- ( -16.89, z-score = -0.85, R) >droPer1.super_6 3352838 92 - 6141320 ----------ACACGCCCACAUUCAAUUACAGUGAAGAUUUGCCGUUUACUUUUCCCCCAGGCC--ACGCCCACAACAUCAAAGGGCGUUGACUUAGACA-UUGA--- ----------...................(((((.......(((................))).--((((((...........)))))).........))-))).--- ( -16.89, z-score = -0.85, R) >droVir3.scaffold_13047 17083968 97 - 19223366 --------UUGCGCAUACGCCGUGUGCUGUACAAUCAAUUACAUGCAAAGCUUUCACUUAGCC---ACGCCCCCAACAUUUAGAGGCGUAACAUUUGGCGCUUGAAGA --------..((((.((((((((((((((((........)))).)))..(((.......))))---))........(.....).)))))).(....)))))....... ( -22.00, z-score = 0.29, R) >droGri2.scaffold_14624 558016 100 + 4233967 ------UUGCGCAAAAGCAACAGCAGCCCGAAAACCAAAUAG--AUUAAUUUAUGGUUCAGUCGGGUUGUUUGUUCAUUAUGAAACCAUAACAACAAAUACAUACACA ------((((......))))..(((((((((.(((((..(((--(....)))))))))...)))))))))(((((..(((((....))))).)))))........... ( -26.00, z-score = -3.89, R) >consensus ____GCAAACACAGGCACACAUUCAAUUACACAUAAGAUUCA__GUUUACUUUCGCCUCGGGC___ACGCCCACAACAUCAAAGGGCGUCAUACU_GACA_UUGA___ ..................................................................((((((...........))))))................... ( -7.39 = -7.27 + -0.12)

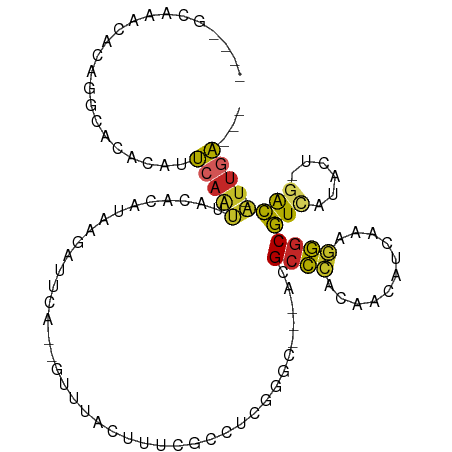
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:43:05 2011