
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,181,709 – 21,181,811 |
| Length | 102 |
| Max. P | 0.922176 |

| Location | 21,181,709 – 21,181,811 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.50 |
| Shannon entropy | 0.58733 |
| G+C content | 0.55558 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -16.63 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21181709 102 - 27905053 ----------------CCGGC--UUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUUUAUGGCACCACUCAGAGCAUCGGUCCCAACGAUCCGAUCUCGGUGCCCAAGCCAAG ----------------..(((--(((..((((((.((((....)))).)))))).............((((((.....(((.(((((((.....)).))))))))))))))))))))... ( -42.20, z-score = -3.18, R) >droGri2.scaffold_14906 5126299 109 + 14172833 ------AAACUAACCACAA-----CUCAGGCCACGCCCUCUUGCGUGAGUGGUCGUUGAUAUUCUAUGGAACAACGCUCACAAUUGAUCCCAAUGAUCCCAUAUCGGUGCCAAGAGCAAG ------.............-----(((.((((((((......))))).(((..(((((...(((....))))))))..)))....((((.....))))..........)))..))).... ( -24.40, z-score = -0.52, R) >droVir3.scaffold_13047 17005555 118 + 19223366 --ACUUUUACUCAUCUCAAAUCCCAUCAGGCCACGCCCUAUUGCGCGAAUGGUCGUUGAUAUUCUAUGGUACCACGCUGACCAUUGAUCCCAAUGAUCCUAUUUCGGUGGCACGUGCUAG --.....................(((...(((((((......)).((((.((((((((....((.(((((.........))))).))...)))))).))...)))))))))..))).... ( -27.80, z-score = -0.79, R) >droMoj3.scaffold_6540 22689282 120 - 34148556 AAAUCGAAAAACCCCCGAACACGAAUCAGGCCAUGCCCUGUUGCGCGAAUGGUCGUUGAUCUUCUAUGGGACCACGCUGACCAUCGAUCCAAAGGAUCCGAUAUCGACGCCGCGUGGCGG ...(((.........)))...........(((((((..(((((..((.((((((((((.(((......))).)).)).)))))).(((((...)))))))....)))))..))))))).. ( -33.10, z-score = 0.50, R) >droWil1.scaffold_181089 5439681 102 + 12369635 ----------------CUAAC--CACUAGGCCAUGCCCUGCUACGGGAAUGGUCAUUGAUAUUCUACGGCACCACGCAAGCCAUCGAUCCCAAUGAUCCGAUCUCAGUGCCGCGCACCAC ----------------.....--.....((((((.(((......))).))))))............((((((..(....)..(((((((.....))).))))....))))))........ ( -28.20, z-score = -1.25, R) >droPer1.super_6 3284474 104 + 6141320 ----------------UCGUCCCCUUUAGGCCAUGCUCUGCUCAGAGAAUGGUCGCUGAUCUUCUACGGCACCACCCUGACCAUCGAUCCCAAUGAUCGGAUUUCGGUGCCGCGUCCCAA ----------------............((((((.((((....)))).))))))............(((((((...........(((((.....)))))(....))))))))........ ( -31.60, z-score = -1.40, R) >dp4.chr2 3256997 104 + 30794189 ----------------UCGUCCUCUUUAGGCCAUGCUCUGCUCAGAGAGUGGUCCCUGAUCUUCUACGGCACCACCCUGACCAUCGAUCCCAAUGAUCGGAUUUCGGUGCCGCGUCCCAA ----------------.........(((((((((.((((....)))).))))..))))).......(((((((...........(((((.....)))))(....))))))))........ ( -31.20, z-score = -1.16, R) >droAna3.scaffold_13340 14409927 104 + 23697760 ----------------CUAAUCUCGACAGGCCAUGCCCUGCUCAGGGAAUGGUCGCUGAUCUUCUACGGCACCACGAACGCCAUCGACCCCAACGACCCCAUCUCGGUGCCGAAGCCGAU ----------------......(((...((((((.((((....)))).))))))((((.....)..(((((((.(((......)))........((.......))))))))).)))))). ( -33.30, z-score = -1.78, R) >droEre2.scaffold_4820 3604730 97 + 10470090 -----------------------UUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUUUAUGGCACCACUCAGAGCAUUGGUCCCAACGAUCCGAUCUCAGUGCCCAGGCCCAG -----------------------..((((((...)).))))...(((..(((.((((((.......(((.((((..........)))).)))..(((...))))))))).)))..))).. ( -29.10, z-score = 0.06, R) >droYak2.chr3R 3520657 104 + 28832112 ----------------CUGGCUUUUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUUUAUGGCACCACUCAGAGCAUUGGUCCCAACGAUCCGAUCUCGGUGCCCAAGCCAAG ----------------.((((((.....((((((.((((....)))).)))))).............((((((.....(((.(((((((.....)).)))))))))))))).)))))).. ( -37.50, z-score = -1.75, R) >droSec1.super_33 435807 102 - 478481 ----------------CCGGC--UUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUUUAUGGCACCACUCAGAGCAUCGGUCCCAACGAUCCGAUCUCGGUGCCCAAGCCAAG ----------------..(((--(((..((((((.((((....)))).)))))).............((((((.....(((.(((((((.....)).))))))))))))))))))))... ( -42.20, z-score = -3.18, R) >droSim1.chr3R 21009135 102 - 27517382 ----------------CCGGC--UUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUUUAUGGCACCACUCAGAGCAUCGGUCCCAACGAUCCGAUCUCGGUGCCCAAGCCAAG ----------------..(((--(((..((((((.((((....)))).)))))).............((((((.....(((.(((((((.....)).))))))))))))))))))))... ( -42.20, z-score = -3.18, R) >consensus ________________CCGGC__CUGUAGGCCAUGCUCUGCUCAGGGAAUGGUCGCUGAUCUUCUAUGGCACCACGCAGACCAUCGAUCCCAACGAUCCGAUCUCGGUGCCCAGGCCAAG ............................((((((.(((......))).)))))).............(((((..........((((.......)))).........)))))......... (-16.63 = -16.67 + 0.04)

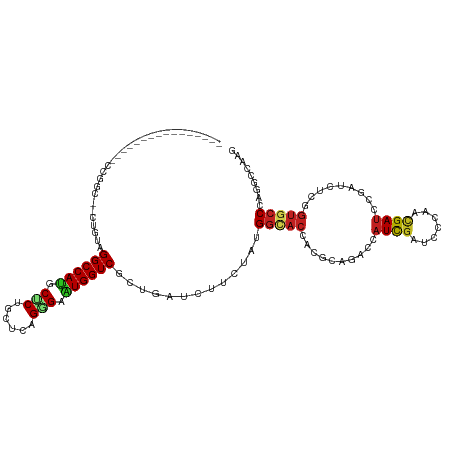
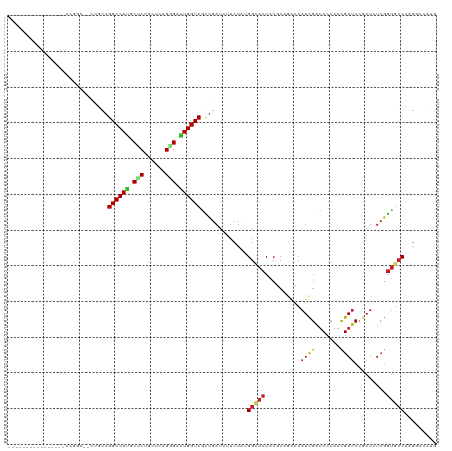
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:59 2011