| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,166,905 – 21,166,998 |
| Length | 93 |
| Max. P | 0.938931 |

| Location | 21,166,905 – 21,166,998 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 61.94 |
| Shannon entropy | 0.82801 |
| G+C content | 0.41931 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -12.78 |
| Energy contribution | -11.91 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.09 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
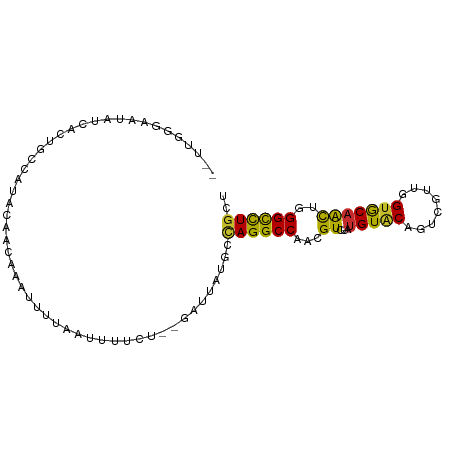
>dm3.chr3R 21166905 93 + 27905053 AUUUGUUAAUAUCAUUGUCAUACAACUAAUUUUAUUUUUCU--GAUUAUGCCAGGCCAACGUCUAUGUACAGUCUUUGGUGCAACUGGGCCUGCU ............(((.((((....................)--))).))).((((((...((...(((((........)))))))..)))))).. ( -16.25, z-score = 0.14, R) >droSim1.chr3R 20993938 93 + 27517382 AUUUGGGAAUAUCAGUGCCACUCAACAAAUUUUAUUUUUCU--GAUUAUGCCAGGCCAACGUCUAUGUACAAUCGUUGGUGCAACUGGGCCUGCU ....(((....(((((..(((.(((((((......)))...--((((.(((.((((....))))..))).)))))))))))..))))).)))... ( -20.20, z-score = 0.10, R) >droSec1.super_33 420619 93 + 478481 AUUUGGGAAUAUCAGUGCCAUACAACUAAUCUUAUUUUUCU--GAUUACACCAGGCCAACGUCUAUGUACAAUCGUUGGUGCAACUGGGCCUGCU ....(((....(((((((........(((((..........--)))))......(((((((............))))))))).))))).)))... ( -19.90, z-score = -0.22, R) >droYak2.chr3R 3505558 95 - 28832112 UCUUGGGAAUAACACUGUCAUACAAGGUAUCUAAUUUCUCUCGGAUUAUGACAGGCCAACGUCUAUGUGCAGUCGUUGGUGCAACUGGGCCUGCU .((..(........((((((((......(((..(......)..)))))))))))(((((((.((......)).)))))))....)..))...... ( -24.20, z-score = -0.45, R) >droEre2.scaffold_4820 3589884 93 - 10470090 --UUUUGGAUGGCACUGCCACAAAAUGCAUCUCAAUUCUCCCUGAUUGUGGCAGGCCAACGUCUAUGUGCAGUCGUUGGUGCAACUGGGCCUGCU --........(((.(((((((((........(((........))))))))))))(((((((.((......)).)))))))........))).... ( -30.40, z-score = -0.78, R) >droAna3.scaffold_13340 14395021 83 - 23697760 -----------UUUCUGCUCUCUGGAGGAUUA-ACUUUCAUGUCCCAACAUUAGGCCAAUGUCUAUGUGCAGUCCUUGGUGCAACUGGGUCUGCU -----------...........((((((....-.)))))).......((((.((((....))))))))((((.(((.(......).))).)))). ( -18.80, z-score = 0.47, R) >droWil1.scaffold_181130 3204122 89 - 16660200 -AAUGAAACCAUCUCCGACCUUAAAUAAUUUAUGAACUUCU-----CUUUAUAGGCCAAUGUGUACGUACAAUCCUUGGUACAAUUGGGUCUGCU -...............(((((........(((((((.....-----.)))))))(((((..(((....)))....)))))......))))).... ( -13.50, z-score = -0.00, R) >droVir3.scaffold_13047 16988222 87 - 19223366 --------AUAUUAAGAUUUUAUACAACAUACUGAAUCCUGUUCUUGUUUUCAGGCCAAUGUUUAUGUGCAGUCGCUGGUACAAUUGGGUUUGCU --------.................(((...(((((..(.......)..))))).(((((.....(((((........))))))))))))).... ( -13.20, z-score = 1.17, R) >droMoj3.scaffold_6540 22671918 89 + 34148556 --ACACUUAAAUUAUAAAACUCGACUAAACACUGAAUCGUA----UAUUCUCAGGCCAAUGUCUAUGUUCAGUCGCUGGUGCAGCUGGGUUUACU --..............((((((((((.((((..((((....----.))))..((((....)))).)))).))))((((...)))).))))))... ( -18.30, z-score = -1.29, R) >consensus __UUGGGAAUAUCACUGCCAUACAACAAAUUUUAAUUUUCU__GAUUAUGCCAGGCCAACGUCUAUGUACAGUCGUUGGUGCAACUGGGCCUGCU ...................................................((((((...((...(((((........)))))))..)))))).. (-12.78 = -11.91 + -0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:58 2011