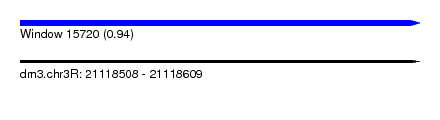
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,118,508 – 21,118,609 |
| Length | 101 |
| Max. P | 0.937796 |

| Location | 21,118,508 – 21,118,609 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Shannon entropy | 0.50223 |
| G+C content | 0.46431 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -13.97 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.937796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
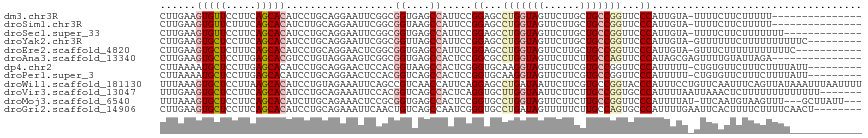
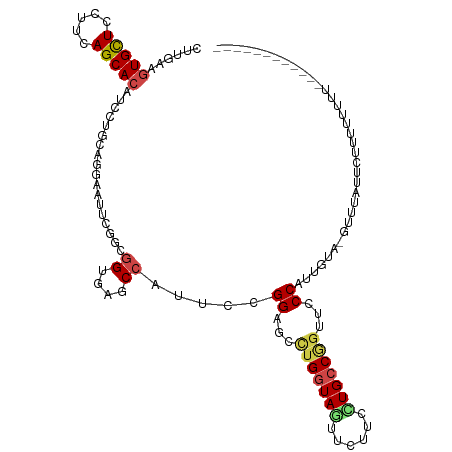
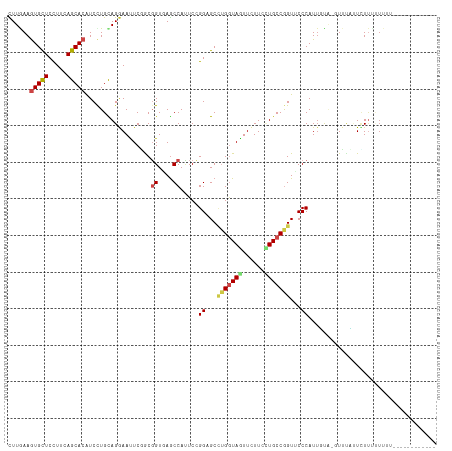
>dm3.chr3R 21118508 101 + 27905053 CUUGAAGUGUUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGUA-UUUUCUUCUUUUU--------------- ...((((.((((((.(((......))).))))))..(((.....))).....((((((.((((((....)))))))))))).......-....)))).....--------------- ( -34.30, z-score = -2.93, R) >droSim1.chr3R 20948984 101 + 27517382 CUUGAAGUGUUCUUUCAGCACAUCUUGCAGGAAUUCGGCGGUAAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGUA-UUUUCUUCUUUUU--------------- ...((.(((((.....))))).)).(((((((((..(((.....))))))))((((((.((((((....))))))))))))...))))-.............--------------- ( -34.20, z-score = -3.64, R) >droSec1.super_33 378261 103 + 478481 CUUGAAGUGUUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGUA-UUUUCUUCUUUUUUU------------- ...((((.((((((.(((......))).))))))..(((.....))).....((((((.((((((....)))))))))))).......-....)))).......------------- ( -34.30, z-score = -2.97, R) >droYak2.chr3R 3461428 107 - 28832112 CUUGAAGUGCUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUUAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGUA-GUUUUUUCUUUUUUUUUUC--------- ...((((((((.....)))))...((((((((((..(((.....))))))))((((((.((((((....))))))))))))...))))-)....)))...........--------- ( -39.10, z-score = -4.83, R) >droEre2.scaffold_4820 3545005 105 - 10470090 CUUGAAGUGCUCUUUCAGCACAUCCUGCAGGAACUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUGCUGCCGGUUCCCAUUGUA-GUUUCUUUUUUUUUUUC----------- ...((((((((.....)))))...(((((((((...(((.....))).))))((((((.((((((....))))))))))))...))))-).)))............----------- ( -37.90, z-score = -3.61, R) >droAna3.scaffold_13340 1313393 102 + 23697760 CUUGAAGUGCUCCUUGAGCACGUCCUGUAGGAAGUCGGCGGUGAGCCACUCCGGCGCCUGGUAGUUCUUCUUGCCAGUUCCCAUAGCGAGUUUUGUAUUAGA--------------- ((((..((((((...))))))...((((.(((((((((.((((...)))))))))..(((((((......))))))))))).))))))))............--------------- ( -34.50, z-score = -1.46, R) >dp4.chr2 10239190 107 - 30794189 CUUAAAAUGCUCCUUGAGCACAUCCUGCAGGAACUCCACGGUAAGCCACUCGGGUGCAAGGUAGUUCUUCGUGCCGGUUCCCAUUUUU-CUGUGUUCUUUCUUUUAUU--------- ...............(((((((.......((((((....((....)).....((..(((((....)))).)..)))))))).......-.)))))))...........--------- ( -25.26, z-score = -0.67, R) >droPer1.super_3 4070128 107 - 7375914 CUUAAAAUGCUCCUUGAGCACAUCCUGCAGGAACUCCACGGUCAGCCACUCCGGUGCAAGGUAGUUCUUCGUGCCGGUUCCCAUUUUU-CUGUGUUCUUUCUUUUAUU--------- ...............(((((((.......(((((.....((....))....(((..(((((....)))).)..)))))))).......-.)))))))...........--------- ( -26.76, z-score = -1.66, R) >droWil1.scaffold_181130 3156685 117 - 16660200 UUUAAAGUGCUCCUUAAGCACAUCCUGUAGAAAUUCAGCCGUCAACCAUUCAGGAGCCUGAUAAUUCUUCGUGCCGGUACCCAUUUCCUGUUCAAUUUCAGUUAUAAAUUUAAUUUU .((((((((((.....)))))....(((((((((...((((.((.....((((....))))..........)).))))....)))))(((........))).))))..))))).... ( -18.56, z-score = -1.09, R) >droVir3.scaffold_13047 16946740 110 - 19223366 UUUGAAGUGCUCCUUCAGCACAUCCUGCAGAAAUUCCACGGUCAGCCACUCAGGUGCUUGGUAAUUCUUCUUGCCGGUGCCCAUUUUAAUUAAACUCUUUUUUUUUUUUU------- ..(((.(((((.....)))))..................((....))..)))((..((.(((((......)))))))..)).............................------- ( -20.40, z-score = -0.23, R) >droMoj3.scaffold_6540 22632632 110 + 34148556 UUUAAAGUGCUCCUUCAGCACAUCUUGCAGAAACUCCGCGGUGAGCCACUCCGGUGCCUGGUAGUUCUUCUUGCCGGUUCCCAUUUUAU-UUCAAUGUAAGUUU---GCUUAUU--- ......(((((.....)))))..(((((((((((.(((.((((...)))).))).).(((((((......)))))))...........)-)))..))))))...---.......--- ( -25.30, z-score = -0.82, R) >droGri2.scaffold_14906 5035436 109 - 14172833 CUUGAAGUGCUCCUUCAGCACAUCCUGCAGAAAUUCAACUGUCAGCCAAUCGGGUGCCUGAUAGUUUUUCUUGCCAGUGCCCAUUUUGAAUUCACUUUUCUUUUCAACU-------- .((((((((((.....)))))...(((((((((...(((((((((.((......)).))))))))))))).)).))).........................)))))..-------- ( -24.40, z-score = -1.15, R) >consensus CUUGAAGUGCUCCUUCAGCACAUCCUGCAGGAAUUCGGCGGUGAGCCAUUCCGGAGCCUGGUAGUUCUUCCUGCCGGUUCCCAUUGUA_GUUUAUUCUUUUUUUU____________ ......(((((.....)))))..................((....)).....((...(((((((......)))))))...))................................... (-13.97 = -14.50 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:57 2011