| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,058,865 – 21,058,964 |
| Length | 99 |
| Max. P | 0.898518 |

| Location | 21,058,865 – 21,058,964 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.85 |
| Shannon entropy | 0.58585 |
| G+C content | 0.51783 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -9.54 |
| Energy contribution | -9.22 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.898518 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
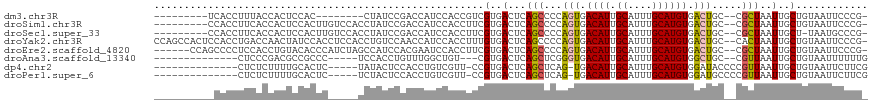
>dm3.chr3R 21058865 99 + 27905053 ---------UCACCUUUACCACUCCAC--------CUAUCCGACCAUCCACCGUCGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC--CGCUAAUUGCUGUAAUUCCCG- ---------......((((........--------...((((((........)))).))...(((..((((.((((.((....)))))).)))).--.))).......))))......- ( -20.20, z-score = -2.79, R) >droSim1.chr3R 20883841 107 + 27517382 ---------CCACCUUCACCACUCCACUUGUCCACCUAUCCGACCAUCCACCUUCGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC--CGCUAAUUGCUGUAAUUCCCG- ---------......((((..........(((.........)))...........))))...(((..((((.((((.((....)))))).)))).--.))).................- ( -17.00, z-score = -1.45, R) >droSec1.super_33 319434 106 + 478481 ---------CCACCUUCACCACUCCACUUGUCCACCUAUCCGACCAUCCACCUUCGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC--CGCUAAUUGCU-UAAUGCCCG- ---------......((((..........(((.........)))...........))))...(((..((((.((((.((....)))))).)))).--.)))....((.-....))...- ( -17.60, z-score = -1.82, R) >droYak2.chr3R 3400385 116 - 28832112 CCAGCCACUCCACCUGACCAACUAUCCACCUCCACCUGUCCAACCAUCCACCUUUGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC--CACUAAUUGCUGUAAUUCCCG- .((((................................((((((..........))).))).......((((.((((.((....)))))).)))).--........)))).........- ( -17.50, z-score = -1.12, R) >droEre2.scaffold_4820 3485591 110 - 10470090 ------CCAGCCCCUCCACCUGUACACCCAUCUAGCCAUCCACGAAUCCACCUUCGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC--CGCUAAUUGCUGUAAUUCCCG- ------.((((.............................((((((......))))))....(((..((((.((((.((....)))))).)))).--.)))....)))).........- ( -25.80, z-score = -3.65, R) >droAna3.scaffold_13340 1251030 95 + 23697760 --------------CUCCCGACGCCGCCC-----UCCACCUGUUUGGCUGU---CGUGACUCAGCUCGGGUGACAUUGCAUUUGCAUGUGGCUGC--CGUUAAUUGCUGUAAUUUUUUG --------------.....(((((.(((.-----..((((((...(((((.---.......)))))))))))((((.((....))))))))).).--)))).................. ( -26.80, z-score = -1.04, R) >dp4.chr2 1451438 98 - 30794189 --------------CUCUCUUUUGCACUC-----UAUACUCCACCUGUCGUU-CCGUGACUCAGCUCAG-UGACAUUGCAUUUGCAUGUGGAUACCCCGUUAAUUGCUGUAAUUCUUCG --------------...............-----............((((..-...)))).((((....-((((..(((....)))...((.....))))))...)))).......... ( -13.50, z-score = 0.51, R) >droPer1.super_6 1430152 98 - 6141320 --------------CUCUCUUUUGCACUC-----UCUACUCCACCUGUCGUU-CCGUGACUCAGCUCAG-UGACAUUGCAUUUGCAUGUGGAUGCCCCGUUAAUUGCUGUAAUUCUUCG --------------...............-----............((((..-...)))).((((....-((((...((((..(....)..))))...))))...)))).......... ( -15.30, z-score = 0.19, R) >consensus __________CACCUUCACCACUCCACUC_____CCUAUCCGACCAUCCACCUUCGUGACUCAGCCCCAGUGACAUUGCAUUUGCAUGUGACUGC__CGCUAAUUGCUGUAAUUCCCG_ .............................................................((((...(((.((((.((....)))))).)))((...)).....)))).......... ( -9.54 = -9.22 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:54 2011