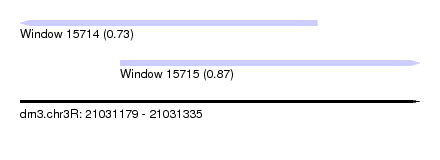
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,031,179 – 21,031,335 |
| Length | 156 |
| Max. P | 0.872380 |

| Location | 21,031,179 – 21,031,295 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.87 |
| Shannon entropy | 0.23100 |
| G+C content | 0.42989 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -21.21 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21031179 116 - 27905053 AUAU-GCGAAUAUUUCACGGACAUUCUCAGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACUGCCAC-CACAAAUUUU-ACCCCAGAUGUUGCAAUCUGGCCCUCGCCCAAAAGG- ....-(((((((((...(((.((((.((....)).))))..)))...)))))))))...............-..........-...((((((......)))))).(((........)))- ( -25.00, z-score = -1.38, R) >droAna3.scaffold_13340 6895149 114 - 23697760 AUAUGGGGAAUAUUUCAUGGACAUUCUCAGUGUACAAUGAACCGAUUGAUAUUCGCUAGAAGAAAUAUAACACUU---UUUUCAGCUGCGAUGUUGCAAUCUGGCCCCAUCCCAAAA--- ..((((((.....(((((..((((.....))))...)))))(((((((....((((..(((((((........))---)))))....)))).....))))).)))))))).......--- ( -30.60, z-score = -2.40, R) >droEre2.scaffold_4820 3456782 119 + 10470090 AUAU-GCGAAUAUUUCACGGACAUUCUCGGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACUACCACUUACAUAUUUUCCCCCCAGAUGUUGCAAUCUGGCCCUCACCCAAAUGGA ....-(((((((((...(((.((((.((....)).))))..)))...)))))))))..............................((((((......)))))).......((....)). ( -25.80, z-score = -1.33, R) >droYak2.chr3R 3372081 112 + 28832112 AUAU-GCGAAUAUUUCACGGACAUUCUCGGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACCAACAC-------UUUUUCCCCCAGAUGUUGCAAUCUGGCCCUCAACCAAAAGGA ....-(((((((((...(((.((((.((....)).))))..)))...)))))))))..((((((.......-------))))))..((((((......)))))).(((........))). ( -27.60, z-score = -2.01, R) >droSec1.super_33 291879 116 - 478481 AUAU-GCGAAUAUUUCACGGACAUUCUCGGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACUGCCAC-CACAAAUUUU-CCCCCAGAUGUUGCAAUCUGGCCCUCACCCAAAAGG- ....-(((((((((...(((.((((.((....)).))))..)))...)))))))))..(((((..((....-..))..))))-)..((((((......)))))).(((........)))- ( -26.50, z-score = -1.53, R) >droSim1.chr3R 20856119 116 - 27517382 AUAU-GCGAAUAUUUCACGGACAUUCUCGGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACUGCCAC-CACAAAUUUU-CCCCCAGAUGUUGCAAUCUGGCCCUCACCCAAAAGG- ....-(((((((((...(((.((((.((....)).))))..)))...)))))))))..(((((..((....-..))..))))-)..((((((......)))))).(((........)))- ( -26.50, z-score = -1.53, R) >consensus AUAU_GCGAAUAUUUCACGGACAUUCUCGGUGGACAAUGAACCGAUUGAUAUUCGCUAGAAAAACUACCAC_CACAAAUUUU_CCCCCAGAUGUUGCAAUCUGGCCCUCACCCAAAAGG_ .....(((((((((...(((.((((.((....)).))))..)))...)))))))))..............................((((((......))))))................ (-21.21 = -21.60 + 0.39)

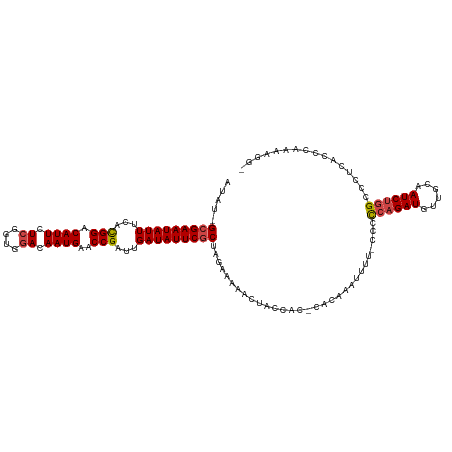
| Location | 21,031,218 – 21,031,335 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.28 |
| Shannon entropy | 0.22375 |
| G+C content | 0.40233 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.87 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21031218 117 + 27905053 -AUUUGUG-GUGGCAGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACUGAGAAUGUCCGUGAAAUAUUCGC-AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGCCAUAAGCA -...(((.-(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))-.........((((((..(((....))).)))))))))))..))) ( -32.80, z-score = -2.44, R) >droAna3.scaffold_13340 6895186 111 + 23697760 ---AAAAGUGUUAUAUUUCUUCUAGCGAAUAUCAAUCGGUUCAUUGUACACUGAGAAUGUCCAUGAAAUAUUCCCCAUAUUGAGUCGAUUUCUUUGCGCCCCAUAAGUCAAGCA------ ---.....................(((((....(((((..(((.(((.....(.((((((.......)))))))..))).)))..)))))..))))).................------ ( -15.30, z-score = 0.53, R) >droEre2.scaffold_4820 3456822 119 - 10470090 AAUAUGUAAGUGGUAGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACCGAGAAUGUCCGUGAAAUAUUCGC-AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGCCAUAAGCA ....(((..(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))-.........((((((..(((....))).)))))))))))..))) ( -31.10, z-score = -2.89, R) >droYak2.chr3R 3372121 112 - 28832112 -------AAGUGUUGGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACCGAGAAUGUCCGUGAAAUAUUCGC-AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGACAUAAGCA -------..(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))-.........((((((..(((....))).)))))))))))..... ( -27.70, z-score = -1.76, R) >droSec1.super_33 291918 117 + 478481 -AUUUGUG-GUGGCAGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACCGAGAAUGUCCGUGAAAUAUUCGC-AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGCCAUAAGCA -...(((.-(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))-.........((((((..(((....))).)))))))))))..))) ( -32.90, z-score = -2.59, R) >droSim1.chr3R 20856158 117 + 27517382 -AUUUGUG-GUGGCAGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACCGAGAAUGUCCGUGAAAUAUUCGC-AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGCCAUAAGCA -...(((.-(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))-.........((((((..(((....))).)))))))))))..))) ( -32.90, z-score = -2.59, R) >consensus _AUUUGUG_GUGGCAGUUUUUCUAGCGAAUAUCAAUCGGUUCAUUGUCCACCGAGAAUGUCCGUGAAAUAUUCGC_AUAUUCAGUUGAUUUCUUGGCGCUCCAUAAGUCAGCCAUAAGCA .........(((((..........((((((((....(((..((((.((....)).)))).)))....))))))))..........((((((..(((....))).)))))))))))..... (-23.14 = -23.87 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:51 2011