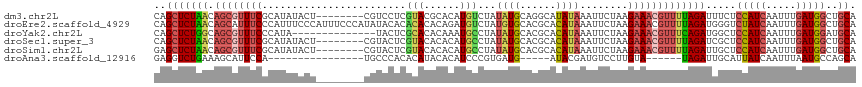
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,591,919 – 8,592,028 |
| Length | 109 |
| Max. P | 0.919889 |

| Location | 8,591,919 – 8,592,028 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.47782 |
| G+C content | 0.41656 |
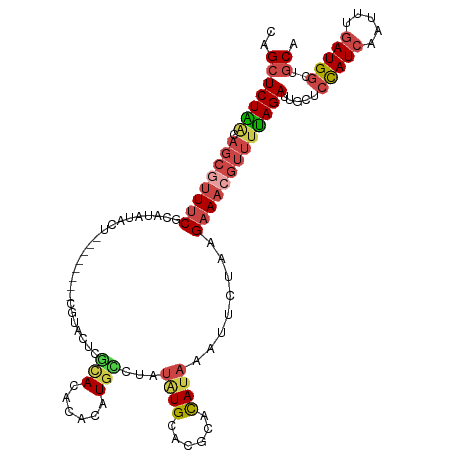
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -13.84 |
| Energy contribution | -15.62 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
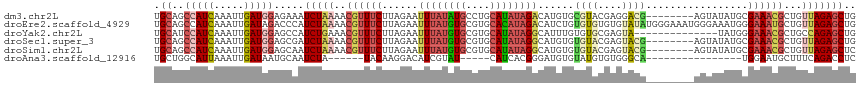
>dm3.chr2L 8591919 109 + 23011544 UGCAGCCAUCAAAUUGAUGGAGAAAUCUAAAACGUUUCUUAGAAUUUAUAUGCCUGCAUAUAGACAUGUGCGUACGAGGACG--------AGUAUAUGCGAAACGCUGUUAGAGCUG .((..(((((.....))))).....(((((..((((((......((((((((....))))))))(((((((...((....))--------.))))))).))))))...))))))).. ( -31.60, z-score = -2.89, R) >droEre2.scaffold_4929 9189093 117 + 26641161 UGCAGCCAUCAAAUUGAUAGACCCAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCACAUAGACAUCUGUGUGUGUGUAUAUGGGAAAUGGGAAAUGGGAAAUGCUGUUAGAGCUG .(((((..((.......((((....))))...(((((((((...((((((((((..(((((((....)))))))..))))))))))...))))))))).))...)))))........ ( -38.70, z-score = -3.76, R) >droYak2.chr2L 11240775 103 + 22324452 UGCAUCCAUCAAAUUGAUGGAGCCAUCUGAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUUUGUGUGCGAGUA--------------UAUGGGAAACGCUGCCAGAGCUG .((.((((((.....))))))))..((((..((((((((.......(((((((.(((((((((....))))))))).)))--------------))))))))))).)..)))).... ( -33.71, z-score = -2.41, R) >droSec1.super_3 4078734 109 + 7220098 UGCAGCCAUCAAAUUGAUGGAGCGAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUACG--------AGUAUAUGCGAAACGCUGUUAGAGCUG .....(((((.....)))))(((..(((((..((((((.((..(((..((..((((((((((....))))))))))..))..--------)))...)).))))))...)))))))). ( -33.80, z-score = -2.09, R) >droSim1.chr2L 8381152 109 + 22036055 UGCAGCCAUCAAAUUGAUGGAGCAAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUACG--------AGUAUAUGCGAAACGCUGUUAGAGCUC (((..(((((.....))))).))).(((((..((((((.((..(((..((..((((((((((....))))))))))..))..--------)))...)).))))))...))))).... ( -34.00, z-score = -2.40, R) >droAna3.scaffold_12916 10237950 90 + 16180835 UGCUGGCAUUAAAUUGAUAAUGCAAUCUA------UACAAGGACAUCGUAU-----CAUCACGGGAUGUGUAUGUGUGGGCA----------------UGGAAUGCUUUCAGACCUC ..(((((((((......))))))...(((------((((.(.(((((.(..-----......).))))).).)))))))(((----------------(...))))...)))..... ( -22.30, z-score = -0.43, R) >consensus UGCAGCCAUCAAAUUGAUGGAGCAAUCUAAAACGUUUCUUAGAAUUUAUGUGCGUGCAUAUAGGCAUGUGUGUACGAGUACG________AGUAUAUGCGAAACGCUGUUAGAGCUG .((..(((((.....))))).....(((((..((((((.......(((((((....)))))))(((....)))..........................))))))...))))))).. (-13.84 = -15.62 + 1.78)
| Location | 8,591,919 – 8,592,028 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.47782 |
| G+C content | 0.41656 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -11.25 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
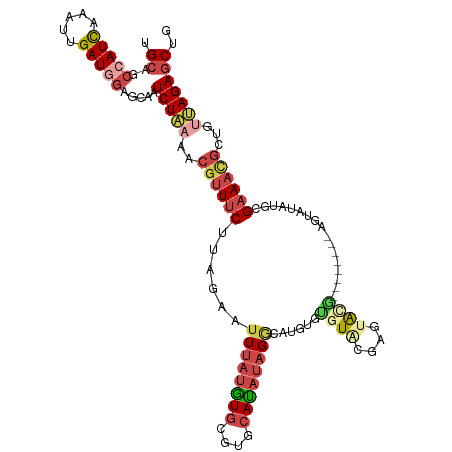
>dm3.chr2L 8591919 109 - 23011544 CAGCUCUAACAGCGUUUCGCAUAUACU--------CGUCCUCGUACGCACAUGUCUAUAUGCAGGCAUAUAAAUUCUAAGAAACGUUUUAGAUUUCUCCAUCAAUUUGAUGGCUGCA (((((((((.((((((((((...(((.--------.......))).))..((((((......))))))...........)))))))))))))......((((.....)))))))).. ( -27.20, z-score = -2.77, R) >droEre2.scaffold_4929 9189093 117 - 26641161 CAGCUCUAACAGCAUUUCCCAUUUCCCAUUUCCCAUAUACACACACACAGAUGUCUAUGUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUGGGUCUAUCAAUUUGAUGGCUGCA .........((((..................((((....................((((((....))))))...(((((((....)))))))))))..((((.....)))))))).. ( -21.50, z-score = -0.74, R) >droYak2.chr2L 11240775 103 - 22324452 CAGCUCUGGCAGCGUUUCCCAUA--------------UACUCGCACACAAAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUCAGAUGGCUCCAUCAAUUUGAUGGAUGCA ....(((((.((((((((.....--------------.....(((......)))...((((......))))........)))))))))))))..((((((((.....)))))).)). ( -27.10, z-score = -2.24, R) >droSec1.super_3 4078734 109 - 7220098 CAGCUCUAACAGCGUUUCGCAUAUACU--------CGUACUCGUACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUCGCUCCAUCAAUUUGAUGGCUGCA ..(.(((((.(((((((((((((((..--------(((............)))..))))))).......((.....)).))))))))))))).)((.(((((.....)))))..)). ( -26.70, z-score = -2.75, R) >droSim1.chr2L 8381152 109 - 22036055 GAGCUCUAACAGCGUUUCGCAUAUACU--------CGUACUCGUACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUUGCUCCAUCAAUUUGAUGGCUGCA ....(((((.(((((((((((((((..--------(((............)))..))))))).......((.....)).))))))))))))).(((.(((((.....)))))..))) ( -25.90, z-score = -2.14, R) >droAna3.scaffold_12916 10237950 90 - 16180835 GAGGUCUGAAAGCAUUCCA----------------UGCCCACACAUACACAUCCCGUGAUG-----AUACGAUGUCCUUGUA------UAGAUUGCAUUAUCAAUUUAAUGCCAGCA ..(((((....((((...)----------------))).....(((.(((.....))))))-----((((((.....)))))------))))))((((((......))))))..... ( -15.20, z-score = 0.39, R) >consensus CAGCUCUAACAGCGUUUCGCAUAUACU________CGUACUCGCACACACAUGCCUAUAUGCACGCACAUAAAUUCUAAGAAACGUUUUAGAUUGCUCCAUCAAUUUGAUGGCUGCA ..(((((((.((((((((........................(((......)))...((((......))))........))))))))))))).....(((((.....)))))..)). (-11.25 = -12.28 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:27 2011