| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,017,671 – 21,017,769 |
| Length | 98 |
| Max. P | 0.838032 |

| Location | 21,017,671 – 21,017,769 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.68516 |
| G+C content | 0.53850 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -13.59 |
| Energy contribution | -13.25 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
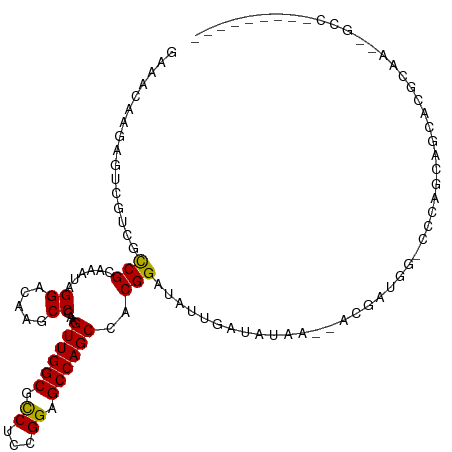
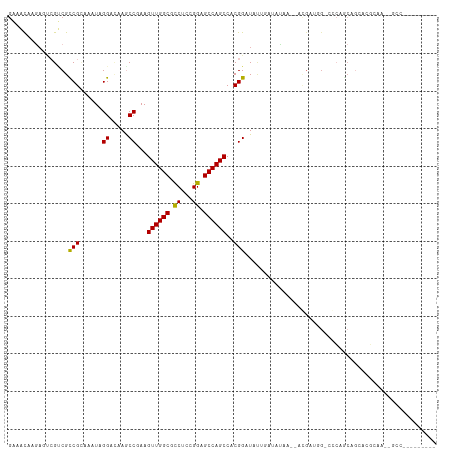
>dm3.chr3R 21017671 98 + 27905053 GGGGCAAGAGUCGCCGUCGCAAAUAGGAAAAGCCAAAGUUGGCGCCCACGGAGCCAGCCACGGAUACAGAGAUAG--AGGAUAGCCCAGACGGUGCUCUG-------------- ......((((.(((((((..............((...((((((.((...)).))))))...))............--.((.....)).))))))))))).-------------- ( -31.50, z-score = -1.03, R) >droEre2.scaffold_4820 3439970 98 - 10470090 GGGGCAAGAGUCGCCGUCGCAAAUAGGAAAAGCCAAAGUUGGCGCCCACGGAGCCAGCCACGGAUACAGAGAUGG--AGGAUAGCCCCAGCGGCGCGCUG-------------- (((((....(((.(((((.(.....((.....))...((((((.((...)).))))))..........).)))))--..))).)))))((((...)))).-------------- ( -34.10, z-score = -0.48, R) >dp4.Unknown_group_4 139024 101 - 150280 GGAACAAGAUCAGACGCCGAAAAUAGGACAAGCCCAAGUUGGCGCCUUCGGAGCCAGCCACGGAUAUUCAUAUAA---CGAAUG-CACAGCAGGAGGCAAUCGCC--------- .......(((.....(((((.....((......))...)))))((((((((......))..(..(((((......---.)))))-..)....)))))).)))...--------- ( -23.80, z-score = -0.32, R) >droPer1.super_3 7179618 101 - 7375914 GGAAUAAGAUCAGACGCCGAAAAUAGGACAAGCCCAAGUUGGCGCCUUCGGAGCCAGCCACGGAUAUUCAUAUAA---CGAAUG-CACAGCAGGAAGCAAUCGCC--------- .(((((.(......).(((......((......))..((((((..(....).))))))..))).)))))......---(((.((-(..........))).)))..--------- ( -22.30, z-score = -0.15, R) >droWil1.scaffold_181130 5786682 89 + 16660200 AAAACAAAAUUCGUCGUCGCAAAUAGGAUAAGCCGAAGUUGGCCUCCUCGGAGCCAGCCACGGAUAAUCAAAUAG--UUGAUG--CCGAGCAA--------------------- .........((((.(((((.......(((...(((..((((((.((....))))))))..)))...)))......--.)))))--.))))...--------------------- ( -25.64, z-score = -2.24, R) >droVir3.scaffold_13047 19197339 110 - 19223366 GAAACAAAAGUCGUCGCCGUAAAUAGGACAAACCGAAGUUGGCACCUCUGGAGCCAGCCACGGAUAUUGUUGAAACUGCGAAGG-AGCACAAGAAUGAAAUGAUCGAUAUA--- .........(((((((((((.....((.....))...((((((.((...)).)))))).)))).((((.(((....(((.....-.)))))).))))...))).))))...--- ( -25.70, z-score = -1.02, R) >droMoj3.scaffold_6540 31804693 113 - 34148556 GAAACAAGUCCCGUCGCCGCAAAUAGGAUAAGCCGAAGUUGGCACCUCUGGAGCCAGCCACGGAUAUUGCUUCAACGGCGAUUG-GGCAGCAGCACGAGAAUGCGACCGAAAUC .......((((.(((((((.....((.(....(((..((((((.((...)).))))))..)))....).))....))))))).)-)))....(((......))).......... ( -38.00, z-score = -1.77, R) >droGri2.scaffold_14906 793925 100 - 14172833 GAAACAAGACGCGUCGCCGCAAAUAGGAUAAACCGAAGUUGGCGCCACUGGAGCCAGCCACGGAUAUAGACACAAUGACAAUGG-CGCAACGUU----GGCUGCC--------- ..........(((((.((.......)).....(((..((((((.((...)).))))))..))).....))).......(((((.-.....))))----)))....--------- ( -27.00, z-score = -0.33, R) >consensus GAAACAAGAGUCGUCGCCGCAAAUAGGACAAGCCGAAGUUGGCGCCUCCGGAGCCAGCCACGGAUAUUGAUAUAA__ACGAUGG_CCCAGCAGCACGCAA__GCC_________ ................(((......((.....))...((((((.((...)).))))))..)))................................................... (-13.59 = -13.25 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:48 2011