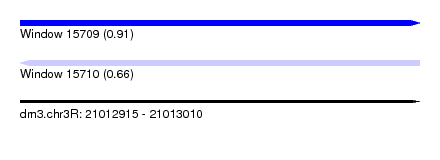
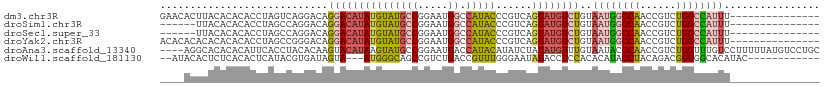
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,012,915 – 21,013,010 |
| Length | 95 |
| Max. P | 0.910110 |

| Location | 21,012,915 – 21,013,010 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.01 |
| Shannon entropy | 0.61913 |
| G+C content | 0.48876 |
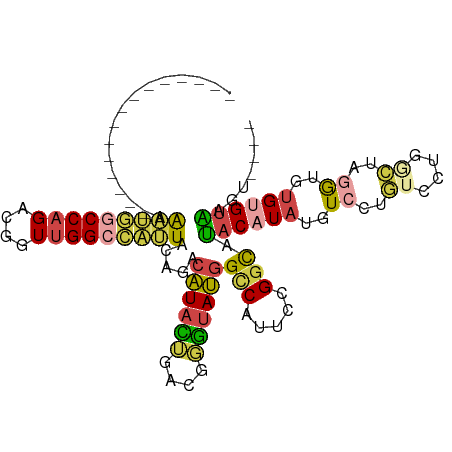
| Mean single sequence MFE | -30.95 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
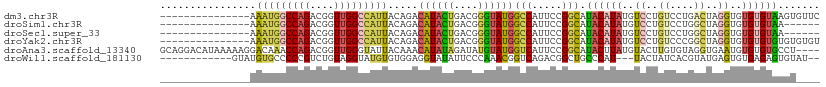
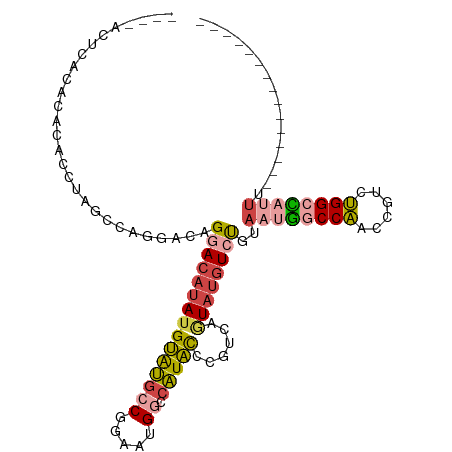
>dm3.chr3R 21012915 95 + 27905053 ---------------AAAUGGCCAGACGGUUGGCCAUUACAGACAUACUGACGGGUAUGGCCAUUCCGGCAUACAUAUGUCCUGUCCUGACUAGGUGUGUGUAAGUGUUC ---------------.(((((((((....))))))))).....((((((....))))))(((.....))).((((((((.(((((....)).)))))))))))....... ( -31.50, z-score = -1.01, R) >droSim1.chr3R 20837807 89 + 27517382 ---------------AAAUGGCCAGACGGUUGGCCAUUACAGACAUACUGACGGGUAUGGCCAUUCCGGCAUACAUAUGUCCUGUCCUGGCUAGGUGUGUGUAA------ ---------------.(((((((((....))))))))).....((((((....))))))(((.....))).((((((((.(((((....)).))))))))))).------ ( -32.30, z-score = -1.32, R) >droSec1.super_33 273671 89 + 478481 ---------------AAAUGGCCAGACGGUUGGCCAUUACAGACAUACUGACGGGUAUGGCCAUUCCGGCAUACAUAUGUCCUGUCCUGGCUAGGUGUGUGUAA------ ---------------.(((((((((....))))))))).....((((((....))))))(((.....))).((((((((.(((((....)).))))))))))).------ ( -32.30, z-score = -1.32, R) >droYak2.chr3R 3352530 95 - 28832112 ---------------AAAUGGCCAGACGGUUGGCCAUUACAGACAUACUGACGGGUAUGGCCAUUCCGGCAUACAUAUGUCCUGUCCCGGCUAGGUGUGUGUGUGUGUGU ---------------.(((((((((....)))))))))(((.((((((.(((((((((((((.....)))...))))...))))))((.....)).)))))).))).... ( -35.80, z-score = -0.98, R) >droAna3.scaffold_13340 6876203 106 + 23697760 GCAGGACAUAAAAAGGACAAACCAGACGGUUGGGUAUUACAAACAUAUAGAUAUGUAUGGUCAUUCCGGCAUACUUAUGUACUUGUGUAGGUGAAUGUGUGUGCCU---- ...(((........((.....)).(((..(((.......))).((((((....)))))))))..)))(((((((((((.(((....))).))))....))))))).---- ( -26.50, z-score = -0.34, R) >droWil1.scaffold_181130 5781620 93 + 16660200 ------------GUAUGUGCCCCCGUCUGUAGGUAUGUGUGGAGGUAUAUUCCCAAACGGUCAGACGGCUGCCCAU---UACUAUCACGUAUGAGUGUGAGAGUGUAU-- ------------..(((.((..(((((((...((...((.((((.....)))))).))...)))))))..)).)))---((((.(((((......))))).))))...-- ( -27.30, z-score = -0.77, R) >consensus _______________AAAUGGCCAGACGGUUGGCCAUUACAGACAUACUGACGGGUAUGGCCAUUCCGGCAUACAUAUGUCCUGUCCUGGCUAGGUGUGUGUAAGU____ ................(((((((((....))))))))).....((((((....))))))(((.....))).((((((..((..((....))..))..))))))....... (-17.84 = -18.27 + 0.42)
| Location | 21,012,915 – 21,013,010 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.01 |
| Shannon entropy | 0.61913 |
| G+C content | 0.48876 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -14.34 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21012915 95 - 27905053 GAACACUUACACACACCUAGUCAGGACAGGACAUAUGUAUGCCGGAAUGGCCAUACCCGUCAGUAUGUCUGUAAUGGCCAACCGUCUGGCCAUUU--------------- ...............(((....)))((((.(((((((((((((.....)).)))))......))))))))))((((((((......)))))))).--------------- ( -26.70, z-score = -0.80, R) >droSim1.chr3R 20837807 89 - 27517382 ------UUACACACACCUAGCCAGGACAGGACAUAUGUAUGCCGGAAUGGCCAUACCCGUCAGUAUGUCUGUAAUGGCCAACCGUCUGGCCAUUU--------------- ------.........(((....)))((((.(((((((((((((.....)).)))))......))))))))))((((((((......)))))))).--------------- ( -26.70, z-score = -1.01, R) >droSec1.super_33 273671 89 - 478481 ------UUACACACACCUAGCCAGGACAGGACAUAUGUAUGCCGGAAUGGCCAUACCCGUCAGUAUGUCUGUAAUGGCCAACCGUCUGGCCAUUU--------------- ------.........(((....)))((((.(((((((((((((.....)).)))))......))))))))))((((((((......)))))))).--------------- ( -26.70, z-score = -1.01, R) >droYak2.chr3R 3352530 95 + 28832112 ACACACACACACACACCUAGCCGGGACAGGACAUAUGUAUGCCGGAAUGGCCAUACCCGUCAGUAUGUCUGUAAUGGCCAACCGUCUGGCCAUUU--------------- ...............(((....)))((((.(((((((((((((.....)).)))))......))))))))))((((((((......)))))))).--------------- ( -26.70, z-score = -0.15, R) >droAna3.scaffold_13340 6876203 106 - 23697760 ----AGGCACACACAUUCACCUACACAAGUACAUAAGUAUGCCGGAAUGACCAUACAUAUCUAUAUGUUUGUAAUACCCAACCGUCUGGUUUGUCCUUUUUAUGUCCUGC ----(((.(((.................((((....))))...(((..(((((.(((((....)))))(((.......))).....)))))..)))......)))))).. ( -14.20, z-score = 1.78, R) >droWil1.scaffold_181130 5781620 93 - 16660200 --AUACACUCUCACACUCAUACGUGAUAGUA---AUGGGCAGCCGUCUGACCGUUUGGGAAUAUACCUCCACACAUACCUACAGACGGGGGCACAUAC------------ --....(((.((((........)))).))).---(((.((..(((((((...((.(((((.......))).))...))...)))))))..)).)))..------------ ( -25.60, z-score = -1.83, R) >consensus ____ACUCACACACACCUAGCCAGGACAGGACAUAUGUAUGCCGGAAUGGCCAUACCCGUCAGUAUGUCUGUAAUGGCCAACCGUCUGGCCAUUU_______________ ............................(((((((((((((((.....)).)))))......))))))))..((((((((......))))))))................ (-14.34 = -15.15 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:47 2011