| Sequence ID | dm3.chr3R |
|---|---|
| Location | 21,005,307 – 21,005,400 |
| Length | 93 |
| Max. P | 0.640287 |

| Location | 21,005,307 – 21,005,400 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 65.26 |
| Shannon entropy | 0.62038 |
| G+C content | 0.39465 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -8.53 |
| Energy contribution | -7.09 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 21005307 93 - 27905053 UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUGCACACACACCCACACACACACAUAUAUAUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ....((((((((((((...(((((((..((.(((((............))))).))..)))))))((....)))))....)))))..)))).. ( -19.40, z-score = -0.37, R) >droSim1.chr3R 20829915 81 - 27517382 UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUG------------CACACACACACACAUAUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ....((((((((((((.(.(((..((.(((.(((((------------......))))).))).))..))).))))....)))))..)))).. ( -17.50, z-score = -0.14, R) >droSec1.super_33 265774 81 - 478481 UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUG------------CACACACACACACAUAUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ....((((((((((((.(.(((..((.(((.(((((------------......))))).))).))..))).))))....)))))..)))).. ( -17.50, z-score = -0.14, R) >droYak2.chr3R 3343345 87 + 28832112 UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUGCACA------CACACACACACAUAUAUAUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ....((((((((((((...(((((((..((.(((((....------..))))).))..)))))))((....)))))....)))))..)))).. ( -20.70, z-score = -1.02, R) >droEre2.scaffold_4820 3427186 79 + 10470090 UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUG--------------CACACACACACUUUUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ...((((((((................))))))))(--------------(((..(((.(.(((((((.....))))))).).)))..)))). ( -18.99, z-score = -1.08, R) >droMoj3.scaffold_6540 31789856 77 + 34148556 ----CGAGUGUGAGGGUGAAACUGCAACCGCAUCUGC-----------AGUGUAUAUACAGAAU-GUAUAUGCACCAUAGCUCGAGGCGCACA ----...(((((.((((...((((((........)))-----------)))((((((((.....-))))))))......))))....))))). ( -26.70, z-score = -2.08, R) >droVir3.scaffold_13047 19183738 77 + 19223366 ----UAUGUAUGCGGCUGCAACCGCAACUGCAUCUGC-----------AGUAUACCAAUACAUA-GUUUAUCCAGCACAACUUGAUAGACACA ----(((((((((((......)))).(((((....))-----------)))......)))))))-(((((((.((.....)).)))))))... ( -25.60, z-score = -4.18, R) >consensus UUUAUACUCAUAUGCCAGGAUAUAUAAAUGAGUGUGC___________CACACACACACACAUAUGCUUAUGCGGCGAAAAUAUGUAAGUGCA ..((((((((((((........))))..)))))))).............................((((((.............))))))... ( -8.53 = -7.09 + -1.44)

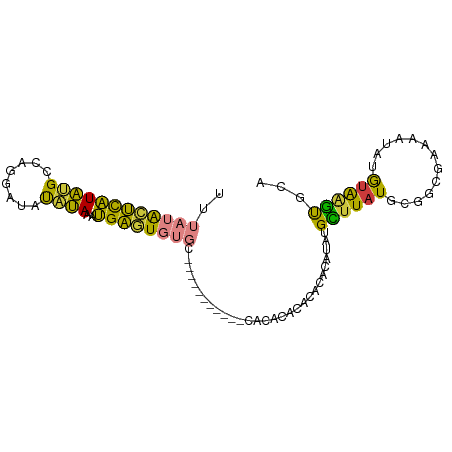
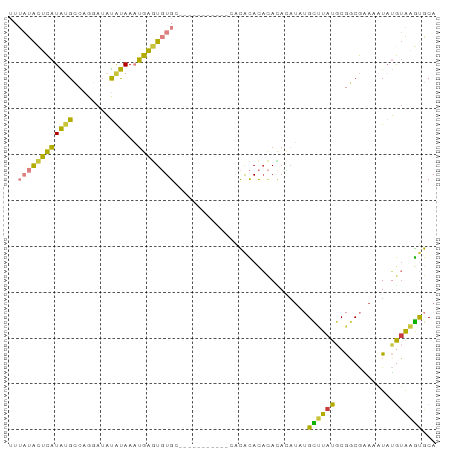
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:45 2011