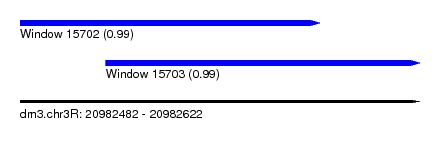
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,982,482 – 20,982,622 |
| Length | 140 |
| Max. P | 0.991292 |

| Location | 20,982,482 – 20,982,587 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.54370 |
| G+C content | 0.36082 |
| Mean single sequence MFE | -18.01 |
| Consensus MFE | -7.57 |
| Energy contribution | -7.65 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
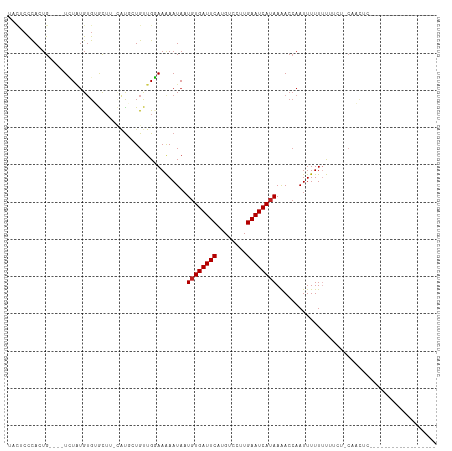
>dm3.chr3R 20982482 105 + 27905053 UACUCCCACUG----UCUAUGUGUGCUU-CAUGCUGUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUUC-CAACUCC---UCCUGCUCGCUUU- ...........----.....(((.((..-...(..((((((((((.(((((((((((......))))))))).........)).))))))-))))..)---....)).)))...- ( -24.20, z-score = -5.32, R) >droSim1.chr3R_random 1022651 99 + 1307089 UACUCCCACUG----UCUAUGUGUGCU--CAUGCUGU-GGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUU----CAACUC----UCCUGCUCGCCUG- ...........----.....(((.((.--.......(-(((((((...(((((((((......))))))))).........))))))----))....----....)).)))...- ( -17.69, z-score = -2.92, R) >droSec1.super_33 243007 103 + 478481 UACUCCCACUG----UCUAUGUGUGCUU-CAUGCUGUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUC---CAACUCC---UCCUGCUUGCCUU- ......(((..----.....))).((..-...(..((((((((((.(((((((((((......)))))))).......))).))))))---))))..)---....)).......- ( -21.61, z-score = -4.39, R) >droYak2.chr3R 3318842 110 - 28832112 UACUCCCACUG----UCUAUGUGUGCUU-CAUGCUGUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUCCCCAACUCCGCCUCCUGCUCGCCUUU ...........----.....(((.((..-...((.((((((((((.(((((((((((......)))))))))........)).)))))))..)))...)).....)).))).... ( -19.90, z-score = -3.43, R) >droEre2.scaffold_4820 3404031 106 - 10470090 UACUCCCACUG----UCUAUGUGUGCUU-CAUGCUGUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGUCCAACUCC---UCCUGCUCGCCUU- ...........----.....(((.((..-...(..((((((.......(((((((((......)))))))))....(((.......)))))))))..)---....)).)))...- ( -22.20, z-score = -4.31, R) >droAna3.scaffold_13340 6844107 99 + 23697760 UACUCCCACUG----UCUA-GUGUAU----GUG-UGCUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGUGCUACUCC---UCCAA--UGCCUU- ......(((..----((((-(..(..----..)-..))))).......(((((((((......)))))))))................))).......---.....--......- ( -17.80, z-score = -2.45, R) >dp4.Unknown_group_4 102378 83 - 150280 UACUCCUAGUG----UUUAUGUGAGCU----UCAUGCUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGCU------------------------ .........((----(((...(.(((.----....))).)..))))).(((((((((......)))))))))...................------------------------ ( -14.10, z-score = -1.23, R) >droPer1.super_3 7140070 81 - 7375914 UACUCCUAGUG----UUUAUGUGAGCU----UCAUGCUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUGCU-------------------------- .........((----(((...(.(((.----....))).)..))))).(((((((((......))))))))).................-------------------------- ( -14.10, z-score = -1.30, R) >droWil1.scaffold_181130 5747640 91 + 16660200 CACUGCUAGUG----UCUGUGUGAGUUU-UGUCCUGCUGGAAAAAUAAUGUGAUUCACGUCCUUGAAUCAGAAGAAAAAUUUUUUUU----CUACGCCUG--------------- (((.....)))----...(((((.((((-(.(((....))))))))....(((((((......)))))))(((((((....))))))----))))))...--------------- ( -17.50, z-score = -1.04, R) >droVir3.scaffold_13047 19148593 92 - 19223366 ----CCUAGUGUACAUAUGUGAGGGUUUUUACCCUGUUGUAAGAAUAAUGUGAUUCCUGUCCUUGAAUCAUAAAACCAAUUUUU-CCACUUCUUUUC------------------ ----...((((((((......(((((....)))))..)))).......((((((((........))))))))............-.)))).......------------------ ( -16.80, z-score = -1.31, R) >droMoj3.scaffold_6540 31757953 88 - 34148556 ----CCUAGUG----UAUUUGUGGGUUUCUAUCCUGUUGUAAGAAUAAUGUGAUUCCUGUCCUUGAAUCAUAAAACCAAUUUUU-CCACUUCUUUUC------------------ ----...((((----........(((((.(((.((......)).))).((((((((........))))))))))))).......-.)))).......------------------ ( -13.79, z-score = -1.30, R) >droGri2.scaffold_14906 13430533 91 + 14172833 --UUCUUAGUG----UAUGUGUGGGUUUUUAUGCUGUUGUAAGAAUAAUGUGAUUCCUGUCCUUGAAUCAUAAAACCAAUUUUUUCCACUUCUUUUC------------------ --.........----.....((((((((((((......))))))))..((((((((........)))))))).............))))........------------------ ( -16.40, z-score = -2.06, R) >consensus UACUCCCACUG____UCUAUGUGUGCUU_CAUGCUGUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUCU_CAACUC__________________ ................................................((((((((........))))))))........................................... ( -7.57 = -7.65 + 0.08)


| Location | 20,982,512 – 20,982,622 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.61 |
| Shannon entropy | 0.26893 |
| G+C content | 0.38043 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.39 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
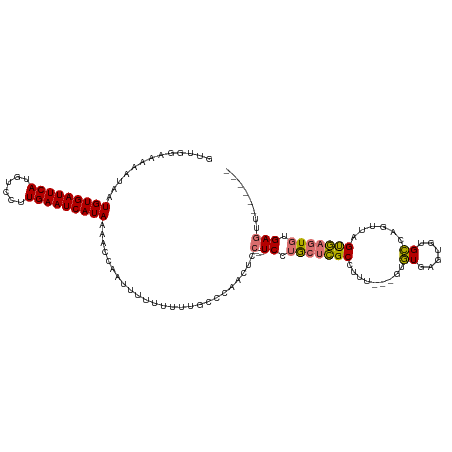
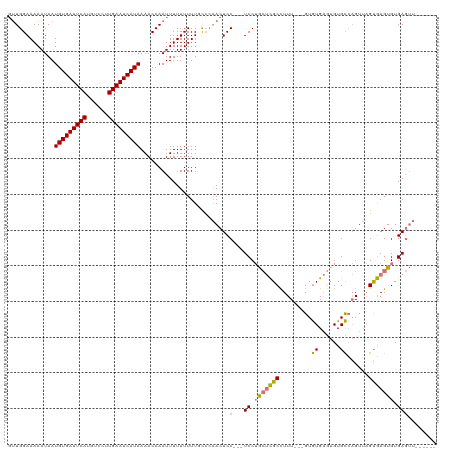
>dm3.chr3R 20982512 110 + 27905053 GUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUUCC-AACUCC---UCCUGCUCGCUUUU---GUGUGAGUGUGCCAGUUAGUGAGUGUGAGUUUUG--- ((((((((((.(((((((((((......))))))))).........)).)))))))-)))..(---((.((((((((.((---(.((......)))))..)))))))).))).....--- ( -32.70, z-score = -5.49, R) >droYak2.chr3R 3318872 120 - 28832112 GUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUCCCCAACUCCGCCUCCUGCUCGCCUUUUGUGUGUGAGUGUGCCAGUUAGUGAGUGUGAGUUUUGGGU (((((........(((((((((......)))))))))...))))).........(((((..((((((((((((((((......).)))))).........)).))).))).)..))))). ( -30.50, z-score = -2.94, R) >droEre2.scaffold_4820 3404061 108 - 10470090 GUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGUCCAACUCC---UCCUGCUCGCCUUU---GUGUGAGUGUGCCAGUUAGUGAGUGUGAGUU------ ((((((.......(((((((((......)))))))))....(((.......)))))))))..(---((.(((((((..((---(.((......)))))...))))))).)))..------ ( -29.40, z-score = -4.45, R) >droAna3.scaffold_13340 6844134 99 + 23697760 --UGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGUGCUACUCC---UCCAA--UGCCUUU---GUAUGUUUGUGU--GUGUGCAAGUGUGA--------- --((((.......(((((((((......)))))))))....(((.......))).........---)))).--.((.(((---(((..(......--)..)))))).))..--------- ( -19.10, z-score = -1.94, R) >consensus GUUGGAAAAAUAAUGUGAUUCAUGUCCUUGAAUCAUAAAACCAAUUUUUUUUUGCCCAACUCC___UCCUGCUCGCCUUU___GUGUGAGUGUGCCAGUUAGUGAGUGUGAGUU______ .............(((((((((......))))))))).............................((.(((((((.......(..(....)..)......))))))).))......... (-14.58 = -14.39 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:41 2011