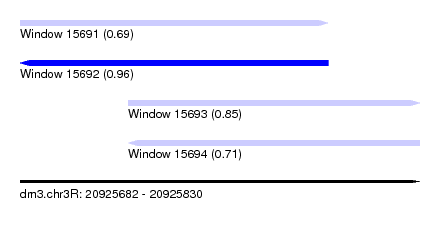
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,925,682 – 20,925,830 |
| Length | 148 |
| Max. P | 0.961573 |

| Location | 20,925,682 – 20,925,796 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.74127 |
| G+C content | 0.40668 |
| Mean single sequence MFE | -23.29 |
| Consensus MFE | -6.48 |
| Energy contribution | -6.56 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20925682 114 + 27905053 GAGUUCUACUAAAGCUCUCGAGCAGAAGUUUCUUUAGCAAAAACAGUCGGGCUCAU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAACUAUCAAGUAAAUC (((((.......)))))..(((.((..(((.....))).......((.(((((...---.......))))).))---.)).))).....(((((((.....)))))))............ ( -26.10, z-score = -2.90, R) >droSim1.chr3R 20755995 114 + 27517382 GAGUUCUGCUAAAGCUCUCGAGCAGAAGUUUCUUUAGCAAAAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAACUAUCAAGUAAAUC (((...((((((((.....((((....))))))))))))......((.(((((...---.......))))).))---....))).....(((((((.....)))))))............ ( -29.80, z-score = -3.20, R) >droYak2.chr3R 3261723 114 - 28832112 GAGUUCUACUAAAGCUCUCGAGCAGAAGUUUCUUUAGCAAAAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAAUUAUCAAGCAAAUC (((((.......)))))..(((.((..(((.....))).......((.(((((...---.......))))).))---.)).))).....(((((((.....)))))))............ ( -23.80, z-score = -1.78, R) >droEre2.scaffold_4820 3345533 114 - 10470090 GAGUUCUACUAAAGCUCUCGAGCAGAAGUUUCUUUAGCAAAAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAACAGGGAACUAUCAAGCAAAUC (((((.......)))))..(((.((..(((.....))).......((.(((((...---.......))))).))---.)).))).....(((((((.....)))))))............ ( -26.10, z-score = -2.91, R) >droAna3.scaffold_13340 6787606 114 + 23697760 GAGUUCUACAAAGGCUCUCGAGCAGAAGUUUCUUUAGCAAAAGCAGUCGGGCUCGU---GUUUUAAAGCCCAAC---CAUUAUCACCACAGUUCUCAAAACUUGAAUUAUUAAAUAAACC (((..((.....((((((...((.((((...)))).))...)).))))(((((...---.......)))))...---............))..)))........................ ( -19.40, z-score = -0.39, R) >droPer1.super_3 7017028 114 - 7375914 GAGUUCUGUUAAAGCUCAUCAACAAAAGUUUCUUUAGCAAAAACAGUCGGGCUCUU---CAUAUGCAGCCCAAA---CAUUGUCACCCGAGCUAUCGAAGACCGAAUUUUCAACUGAAUC ..((((.(((..(((((....((((..(((.....)))..........((((((..---.....).)))))...---..)))).....)))))...(((((.....)))))))).)))). ( -22.00, z-score = -0.97, R) >droWil1.scaffold_181130 11042901 117 - 16660200 GACUUCAACCAAACCCCUCGCAGAGAAGUUUCUUUAGCAAAAAAUUAUGGGCUUUC---UAUCCACAGCCCAAGAAGCCUCCUCCGCGGAAUUAUCAAGUAUAGGUAUUUCAAAAAAUAU ..................(((.(((..((((((..............((((((...---.......))))))))))))...))).)))(((.((((.......)))).)))......... ( -22.63, z-score = -1.53, R) >droVir3.scaffold_13047 314708 118 + 19223366 GACUUCAGUAAAAGAUGUUGAAGAAAAAUUUCUAUAGCAAAAAAAAACGGGCGUGUCUACCAUUUCGCCCAUAAACAAGUGUAAUUUGCAAGUAUCAAAUACACAUUCUUUGAAGGGA-- ..(((((....(((((((((.((((....)))).))))).........(((((............)))))........(((((.((((.......)))))))))..)))))))))...-- ( -25.30, z-score = -2.04, R) >droMoj3.scaffold_6540 1232185 118 + 34148556 GACUACGUCAAAAAUUGUUGCAGAAAAGUUUCUCUAGCAAAAAAAAUCGGGCUACACCGCUAUCAUGGCCCAAAGUAUGUGUUCUUCACAACAAUCAGAUAUUACCUUUUCAAAUGAA-- .......(((....((((((.((((....)))).)))))).....(((((((((...........))))))...((.((((.....)))))).....)))..............))).-- ( -20.50, z-score = -0.92, R) >droGri2.scaffold_14906 446912 115 - 14172833 GACAUCAAUUAAAGAUCUGGCCGAAAAGUUUCUCUAGCAAAAAAAAUUGGGCCUA---ACCUCUUCGGCCCAAAAAAAGUGUUCUUUGCAACUAUCAAAUAUCAGUUAUUCAAAGCGA-- (((((...........((((..(((....))).)))).........(((((((..---........))))))).....)))))(((((.((((..........))))...)))))...-- ( -17.30, z-score = 0.15, R) >consensus GAGUUCUACUAAAGCUCUCGAGCAGAAGUUUCUUUAGCAAAAACAGUCGGGCUCGU___AUUCAAAAGCCCAAC___CCUUCUCACCACAGUUAUCAAACAGGGAAUUAUCAAAUAAAUC ................................................(((((.............)))))................................................. ( -6.48 = -6.56 + 0.08)

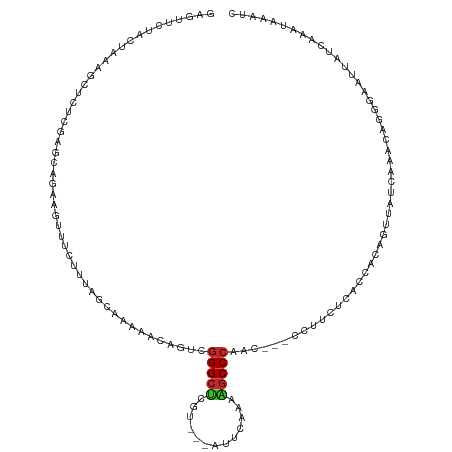
| Location | 20,925,682 – 20,925,796 |
|---|---|
| Length | 114 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.02 |
| Shannon entropy | 0.74127 |
| G+C content | 0.40668 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -9.15 |
| Energy contribution | -9.26 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20925682 114 - 27905053 GAUUUACUUGAUAGUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---AUGAGCCCGACUGUUUUUGCUAAAGAAACUUCUGCUCGAGAGCUUUAGUAGAACUC ......(((.(((((((((.....)))))))))((..((((.(---(((((((((......---..)))))))))).))))..)).)))....(((((((.((....)).)))))))... ( -45.60, z-score = -5.56, R) >droSim1.chr3R 20755995 114 - 27517382 GAUUUACUUGAUAGUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUUUUGCUAAAGAAACUUCUGCUCGAGAGCUUUAGCAGAACUC ......(((.(((((((((.....)))))))))((..((((.(---(((((((((......---..)))))))))).))))..)).)))....(((((((.((....)).)))))))... ( -46.90, z-score = -5.47, R) >droYak2.chr3R 3261723 114 + 28832112 GAUUUGCUUGAUAAUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUUUUGCUAAAGAAACUUCUGCUCGAGAGCUUUAGUAGAACUC ..............(((((.....)))))((.(((..((((.(---(((((((((......---..)))))))))).))))..))).))....(((((((.((....)).)))))))... ( -40.00, z-score = -3.82, R) >droEre2.scaffold_4820 3345533 114 + 10470090 GAUUUGCUUGAUAGUUCCCUGUUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUUUUGCUAAAGAAACUUCUGCUCGAGAGCUUUAGUAGAACUC ......(((.(((((((((.....)))))))))((..((((.(---(((((((((......---..)))))))))).))))..)).)))....(((((((.((....)).)))))))... ( -44.60, z-score = -5.00, R) >droAna3.scaffold_13340 6787606 114 - 23697760 GGUUUAUUUAAUAAUUCAAGUUUUGAGAACUGUGGUGAUAAUG---GUUGGGCUUUAAAAC---ACGAGCCCGACUGCUUUUGCUAAAGAAACUUCUGCUCGAGAGCCUUUGUAGAACUC ..................(((((((((((((.(((..(.(..(---(((((((((......---..))))))))))..).)..))).))....))))(((....))).....))))))). ( -27.40, z-score = -1.02, R) >droPer1.super_3 7017028 114 + 7375914 GAUUCAGUUGAAAAUUCGGUCUUCGAUAGCUCGGGUGACAAUG---UUUGGGCUGCAUAUG---AAGAGCCCGACUGUUUUUGCUAAAGAAACUUUUGUUGAUGAGCUUUAACAGAACUC .....((((......((((((((((.(((((((..(......)---..)))))))....))---))))..))))(((((...(((.....(((....)))....)))...))))))))). ( -29.00, z-score = -0.87, R) >droWil1.scaffold_181130 11042901 117 + 16660200 AUAUUUUUUGAAAUACCUAUACUUGAUAAUUCCGCGGAGGAGGCUUCUUGGGCUGUGGAUA---GAAAGCCCAUAAUUUUUUGCUAAAGAAACUUCUCUGCGAGGGGUUUGGUUGAAGUC ...(((..(.(((..(((......((....))((((((((((......((((((.......---...))))))...((((((....)))))))))))))))))))..))).)..)))... ( -32.20, z-score = -1.39, R) >droVir3.scaffold_13047 314708 118 - 19223366 --UCCCUUCAAAGAAUGUGUAUUUGAUACUUGCAAAUUACACUUGUUUAUGGGCGAAAUGGUAGACACGCCCGUUUUUUUUUGCUAUAGAAAUUUUUCUUCAACAUCUUUUACUGAAGUC --...(((((((((..(((((((((.......)))).))))).((((.(((((((...((.....))))))))).............((((....))))..))))))))....))))).. ( -21.70, z-score = -0.74, R) >droMoj3.scaffold_6540 1232185 118 - 34148556 --UUCAUUUGAAAAGGUAAUAUCUGAUUGUUGUGAAGAACACAUACUUUGGGCCAUGAUAGCGGUGUAGCCCGAUUUUUUUUGCUAGAGAAACUUUUCUGCAACAAUUUUUGACGUAGUC --...((((....)))).(..((.((((((((((((((.........(((((((((.......)))..))))))(((((((....))))))).))))).)))))))))...))..).... ( -22.00, z-score = 0.55, R) >droGri2.scaffold_14906 446912 115 + 14172833 --UCGCUUUGAAUAACUGAUAUUUGAUAGUUGCAAAGAACACUUUUUUUGGGCCGAAGAGGU---UAGGCCCAAUUUUUUUUGCUAGAGAAACUUUUCGGCCAGAUCUUUAAUUGAUGUC --...(((((..((((((........))))))))))).............((((((((((.(---(((...(((......))))))).....))))))))))..(((.......)))... ( -24.00, z-score = -0.21, R) >consensus GAUUUAUUUGAAAAUUCCAUAUUUGAGAACUGUGGUGAGAAGG___GUUGGGCUUUAGAAG___ACGAGCCCGACUGUUUUUGCUAAAGAAACUUCUGCUCGAGAGCUUUAGUAGAACUC ........................(((....................(((((((.............)))))))..(((((((....(((....)))...)))))))..........))) ( -9.15 = -9.26 + 0.11)



| Location | 20,925,722 – 20,925,830 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 59.30 |
| Shannon entropy | 0.83687 |
| G+C content | 0.39322 |
| Mean single sequence MFE | -20.24 |
| Consensus MFE | -6.48 |
| Energy contribution | -6.56 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.852875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20925722 108 + 27905053 AAACAGUCGGGCUCAU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAACUAUCAAGUAAAUCGUGCGGAAACAGACA-UAGUAAA---CAAAAUAGGAAA .....((((((((...---.......)))))...---.....((..((((((((((.....))))))).....(.....))))..))....))).-.......---............ ( -25.10, z-score = -2.99, R) >droSim1.chr3R 20756035 108 + 27517382 AAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAACUAUCAAGUAAAUCGUGCGGAAACAGACA-UAGUAAA---CAAAAUAGGAAA .....((((((((...---.......)))))...---.....((..((((((((((.....))))))).....(.....))))..))....))).-.......---............ ( -25.10, z-score = -2.62, R) >droYak2.chr3R 3261763 108 - 28832112 AAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAGCAGGGAAUUAUCAAGCAAAUCGUGCGGAAACAGACA-UAGCAAA---CAAAAUAGGAAA .....((((((((...---.......)))))...---........((..(((((((.....))))))).....(((.....))))).....))).-.......---............ ( -24.30, z-score = -2.33, R) >droEre2.scaffold_4820 3345573 108 - 10470090 AAACAGUCGGGCUCGU---AUUCAAAAGCCCAAC---CCUUCUCACCACAGUUCCCAAACAGGGAACUAUCAAGCAAAUCGUGCGGAAACAGACA-UAGCAAA---CAAAAUAGGAAA .....((((((((...---.......)))))...---........((..(((((((.....))))))).....(((.....))))).....))).-.......---............ ( -26.60, z-score = -3.42, R) >droAna3.scaffold_13340 6787646 108 + 23697760 AAGCAGUCGGGCUCGU---GUUUUAAAGCCCAAC---CAUUAUCACCACAGUUCUCAAAACUUGAAUUAUUAAAUAAACCGUGCGGUAACAGACA-GAACAAA---CAACAUAGAAAA .....((((((((...---.......)))))...---............(((((.........))))).........(((....)))....))).-.......---............ ( -15.00, z-score = 0.16, R) >droPer1.super_3 7017068 108 - 7375914 AAACAGUCGGGCUCUU---CAUAUGCAGCCCAAA---CAUUGUCACCCGAGCUAUCGAAGACCGAAUUUUCAACUGAAUCAUGCGUAAAUGGAUA-CCAAAAA---CAAUCUAAAAAA ...((((.((((((..---.....).)))))...---....(((...(((....)))..)))..........))))..((((......))))...-.......---............ ( -16.80, z-score = -0.14, R) >droWil1.scaffold_181130 11042941 111 - 16660200 AAAAUUAUGGGCUUUC---UAUCCACAGCCCAAGAAGCCUCCUCCGCGGAAUUAUCAAGUAUAGGUAUUUCAAAAAAUAUGUGCGAAAAUAGUAA-UAAUAGA---CAGAUAGGAAAA .......((((((...---.......)))))).....(((((.....)))..((((..((((..((((((....)))))))))).......((..-......)---).)))))).... ( -16.60, z-score = 0.56, R) >droVir3.scaffold_13047 314748 111 + 19223366 AAAAAAACGGGCGUGUCUACCAUUUCGCCCAUAAACAAGUGUAAUUUGCAAGUAUCAAAUACACAUUCUUUGAAGGGA-UACAAAAACACUGA---UAACCGA---CAACUUGGAUAU ........(((((............))))).......(((((..((((...((((...))))..(((((.....))))-).)))).)))))..---...((((---....)))).... ( -18.60, z-score = -0.41, R) >droMoj3.scaffold_6540 1232225 111 + 34148556 AAAAAAUCGGGCUACACCGCUAUCAUGGCCCAAAGUAUGUGUUCUUCACAACAAUCAGAUAUUACCUUUUCAAAUGAA-UAUGAAAAAAUUGACUCUAA--AA----AACUCGAAAAU .....(((((((((...........))))))...((.((((.....)))))).....)))......((((((......-..))))))............--..----........... ( -15.70, z-score = -1.13, R) >droGri2.scaffold_14906 446952 114 - 14172833 AAAAAAUUGGGCCUA---ACCUCUUCGGCCCAAAAAAAGUGUUCUUUGCAACUAUCAAAUAUCAGUUAUUCAAAGCGA-CAUGAAAACAUAGAAACCAGCCGAAACCAACCUGAAAAU ......(((((((..---........))))))).....((((((((((.((((..........))))...))))).))-))).................................... ( -18.60, z-score = -1.28, R) >consensus AAACAGUCGGGCUCGU___AUUCAAAAGCCCAAC___CCUUCUCACCACAGUUAUCAAACAGGGAAUUAUCAAAUAAAUCGUGCGGAAACAGACA_UAACAAA___CAAAAUAGAAAA ........(((((.............)))))....................................................................................... ( -6.48 = -6.56 + 0.08)



| Location | 20,925,722 – 20,925,830 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 59.30 |
| Shannon entropy | 0.83687 |
| G+C content | 0.39322 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -7.09 |
| Energy contribution | -7.51 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20925722 108 - 27905053 UUUCCUAUUUUG---UUUACUA-UGUCUGUUUCCGCACGAUUUACUUGAUAGUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---AUGAGCCCGACUGUUU ............---.(((((.-.((((((....))).))).......(((((((((.....))))))))))))))(((.(---(((((((((......---..)))))))))).))) ( -35.50, z-score = -3.84, R) >droSim1.chr3R 20756035 108 - 27517382 UUUCCUAUUUUG---UUUACUA-UGUCUGUUUCCGCACGAUUUACUUGAUAGUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUU ............---.(((((.-.((((((....))).))).......(((((((((.....))))))))))))))(((.(---(((((((((......---..)))))))))).))) ( -34.80, z-score = -3.40, R) >droYak2.chr3R 3261763 108 + 28832112 UUUCCUAUUUUG---UUUGCUA-UGUCUGUUUCCGCACGAUUUGCUUGAUAAUUCCCUGCUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUU ..((((((.(((---((.((..-.((((((....))).)))..))..)))))(((((.....)))))..)))).))(((.(---(((((((((......---..)))))))))).))) ( -31.90, z-score = -2.39, R) >droEre2.scaffold_4820 3345573 108 + 10470090 UUUCCUAUUUUG---UUUGCUA-UGUCUGUUUCCGCACGAUUUGCUUGAUAGUUCCCUGUUUGGGAACUGUGGUGAGAAGG---GUUGGGCUUUUGAAU---ACGAGCCCGACUGUUU ...(((.(((..---((.((..-.((((((....))).)))..))...(((((((((.....)))))))))))..))))))---(((((((((......---..)))))))))..... ( -35.80, z-score = -3.54, R) >droAna3.scaffold_13340 6787646 108 - 23697760 UUUUCUAUGUUG---UUUGUUC-UGUCUGUUACCGCACGGUUUAUUUAAUAAUUCAAGUUUUGAGAACUGUGGUGAUAAUG---GUUGGGCUUUAAAAC---ACGAGCCCGACUGCUU ............---.......-....((((((((((..((((.(..(((.......)))..).))))))))))))))..(---(((((((((......---..)))))))))).... ( -27.10, z-score = -1.74, R) >droPer1.super_3 7017068 108 + 7375914 UUUUUUAGAUUG---UUUUUGG-UAUCCAUUUACGCAUGAUUCAGUUGAAAAUUCGGUCUUCGAUAGCUCGGGUGACAAUG---UUUGGGCUGCAUAUG---AAGAGCCCGACUGUUU ..((((.(((((---..(((((-((......))).)).))..))))).)))).((((((((((.(((((((..(......)---..)))))))....))---))))..))))...... ( -23.00, z-score = 0.45, R) >droWil1.scaffold_181130 11042941 111 + 16660200 UUUUCCUAUCUG---UCUAUUA-UUACUAUUUUCGCACAUAUUUUUUGAAAUACCUAUACUUGAUAAUUCCGCGGAGGAGGCUUCUUGGGCUGUGGAUA---GAAAGCCCAUAAUUUU .....((.((((---(((((..-......(((((((...(((((....))))).........((....)).))))))).((((.....)))))))))))---)).))........... ( -19.00, z-score = 0.38, R) >droVir3.scaffold_13047 314748 111 - 19223366 AUAUCCAAGUUG---UCGGUUA---UCAGUGUUUUUGUA-UCCCUUCAAAGAAUGUGUAUUUGAUACUUGCAAAUUACACUUGUUUAUGGGCGAAAUGGUAGACACGCCCGUUUUUUU ....(((..(((---((.((.(---(.(((((.((((((-.....(((((.........)))))....))))))..))))).))..)).)))))..)))................... ( -22.10, z-score = -0.35, R) >droMoj3.scaffold_6540 1232225 111 - 34148556 AUUUUCGAGUU----UU--UUAGAGUCAAUUUUUUCAUA-UUCAUUUGAAAAGGUAAUAUCUGAUUGUUGUGAAGAACACAUACUUUGGGCCAUGAUAGCGGUGUAGCCCGAUUUUUU .((((((....----..--(((((....(((((((((..-......)))))))))....)))))......)))))).........(((((((((.......)))..))))))...... ( -22.40, z-score = -0.83, R) >droGri2.scaffold_14906 446952 114 + 14172833 AUUUUCAGGUUGGUUUCGGCUGGUUUCUAUGUUUUCAUG-UCGCUUUGAAUAACUGAUAUUUGAUAGUUGCAAAGAACACUUUUUUUGGGCCGAAGAGGU---UAGGCCCAAUUUUUU .......((((..((((((((.(......((((((..((-(.((((..((((.....))))..).))).))).)))))).......).))))))))....---..))))......... ( -24.32, z-score = -0.06, R) >consensus UUUUCUAUUUUG___UUUGCUA_UGUCUGUUUCCGCACGAUUUAUUUGAAAAUUCCAUAUUUGAGAACUGUGGUGAGAAGG___GUUGGGCUUUAGAAG___ACGAGCCCGACUGUUU .....................................................................................(((((((.............)))))))...... ( -7.09 = -7.51 + 0.42)

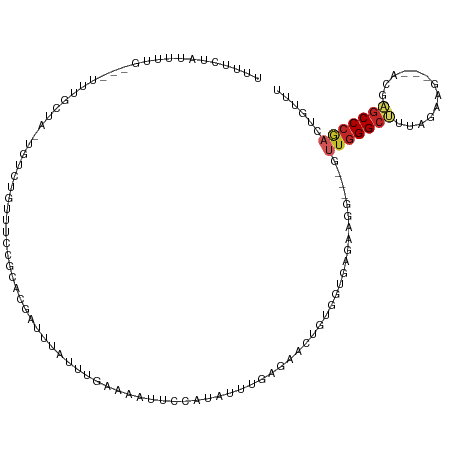
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:33 2011