| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,586,294 – 8,586,386 |
| Length | 92 |
| Max. P | 0.913334 |

| Location | 8,586,294 – 8,586,386 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.71 |
| Shannon entropy | 0.65388 |
| G+C content | 0.51547 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -8.17 |
| Energy contribution | -8.37 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
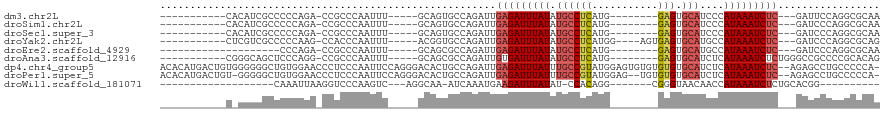
>dm3.chr2L 8586294 92 - 23011544 -----------CACAUCGCCCCCAGA-CCGCCCAAUUU-----GCAGUGCCAGAUUGAGAUUUAUAUGCCUCAUG--------GAGUGCAUCCCAUAAAUCUC---GAUUCCAGGCGCAA -----------...............-..((.......-----)).(((((.((((((((((((((((((((...--------))).))))...)))))))))---))))...))))).. ( -27.40, z-score = -3.25, R) >droSim1.chr2L 8375590 92 - 22036055 -----------CACAUCGCCCCCAGA-CCGCCCAAUUU-----GCAGUGCCAGAUUGAGAUUUAUAUGCCUCAUG--------GAGUGCAUCCCAUAAAUCUC---GAUCCCAGGCGCAA -----------...............-..((.......-----)).(((((.((((((((((((((((((((...--------))).))))...)))))))))---))))...))))).. ( -29.20, z-score = -3.95, R) >droSec1.super_3 4073195 92 - 7220098 -----------CACAUCGCCCCCAGA-CCGCCCAAUUU-----GCAGUGCCAGAUUGAGAUUUAUAUGCCUCAUG--------GAGUGCAUCCCAUAAAUCUC---GAUCCCAGGCGCAA -----------...............-..((.......-----)).(((((.((((((((((((((((((((...--------))).))))...)))))))))---))))...))))).. ( -29.20, z-score = -3.95, R) >droYak2.chr2L 11234502 96 - 22324452 -----------CUCGUCGCCCCCAAG-CCACCCAAUUU-----ACGGUGCCAGAUUGAGAUUUAUAUGCCUCAUGG----AGUGAGUGCAUGCCAUAAAUCUC---GAUCCCAGGCGCAG -----------...(.((((.....(-.((((......-----..)))).).((((((((((((((((((((((..----.))))).))))...)))))))))---))))...))))).. ( -34.60, z-score = -4.40, R) >droEre2.scaffold_4929 9182515 83 - 26641161 --------------------CCCAGA-CCGCCCAAUUU-----GCAGCGCCAGAUUGAGAUUUAUAUGCCUCAUG--------GAGUGCAUGCCAUAAAUCUC---GAUCCCAGGCGCAA --------------------......-..((.......-----)).(((((.((((((((((((((((((((...--------))).))))...)))))))))---))))...))))).. ( -31.10, z-score = -4.60, R) >droAna3.scaffold_12916 10231411 95 - 16180835 -----------CGGGCAGCUCCCAGG-CCGCCCAAUUU-----GCAGCGCCAGAUUGUGAUUUAUAUGCCUCAUG--------GAGUGCAUCUCAUAAAUCUCUGGGCCGCCCCGCACAG -----------.((((.(((....))-).)))).....-----((.((.(((((....((((((((((((((...--------))).))))...)))))))))))))).))......... ( -32.60, z-score = -1.03, R) >dp4.chr4_group5 1344786 117 + 2436548 ACACAUGACUGUGGGGGGCUGUGGAACCCUCCCAAUUCCAGGGACACUGCCAGAUUGAGAUUUAUUUGCCGUAUGGAGUGUGUGUGUGCAUCUCAUAAAUCUC--AGAGCCUGCCCCCA- .((((....))))(((((((.(((.....((((.......)))).....))).)(((((((((((.((((((((.......))))).)))....)))))))))--)).....)))))).- ( -40.10, z-score = -0.77, R) >droPer1.super_5 1315464 114 + 6813705 ACACAUGACUGU-GGGGGCUGUGGAACCCUCCCAAUUCCAGGGACACUGCCAGAUUGAGAUUUAUUUGCCGUAUGGAG--UGUGUGUGCAUCUCAUAAAUCUC--AGAGCCUGCCCCCA- ...........(-(((((((.(((.....((((.......)))).....))).)(((((((((((.((((((((....--.))))).)))....)))))))))--)).....)))))))- ( -37.60, z-score = -0.77, R) >droWil1.scaffold_181071 1006553 79 - 1211509 -------------------CAAAUUAAGGUCCCAAGUC---AGGCAA-AUCAAAUGAAGAUUUAUAU-CCACAGG-------CGGGUAACAACCAUAAAUCUCUGCACGG---------- -------------------................(((---((....-.((....))((((((((((-((.....-------.))))).......)))))))))).))..---------- ( -9.51, z-score = 0.77, R) >consensus ___________C_CGUCGCCCCCAGA_CCGCCCAAUUU_____GCAGUGCCAGAUUGAGAUUUAUAUGCCUCAUG________GAGUGCAUCCCAUAAAUCUC___GAUCCCAGGCGCAA ........................................................(((((((((.((((((...........))).)))....)))))))))................. ( -8.17 = -8.37 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:25 2011