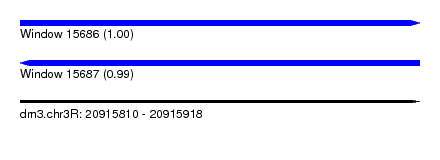
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,915,810 – 20,915,918 |
| Length | 108 |
| Max. P | 0.997701 |

| Location | 20,915,810 – 20,915,918 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.86 |
| Shannon entropy | 0.12947 |
| G+C content | 0.48158 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -34.30 |
| Energy contribution | -35.13 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
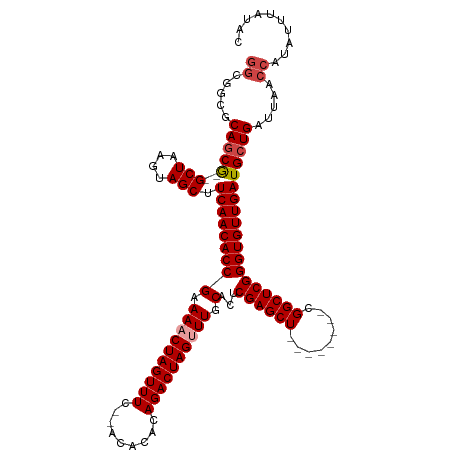
>dm3.chr3R 20915810 108 + 27905053 GGCGGCGCAGCG--GCUAAGUAGCUUCAACACCGAAAACUAGUUUC--ACACACAGACUAGUUUGCACUCGAGCU--------CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC .((......))(--(.(((.((((.(((((((((.((((((((((.--......)))))))))).)...(((((.--------..))))))))))))).)))).))).)).......... ( -39.00, z-score = -3.95, R) >droYak2.chr3R 3251328 118 - 28832112 GGCAUCGCAGCGAAGCUAAGUAGCUUCAACACCGAAAACUAGUUUC--ACACACAGACUAGUUUGCACUCGAGCUUGGGUGCUCGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC ((((((.....((((((....))))))(((((((.((((((((((.--......)))))))))).)..((((((......))))))....)))))))))))).................. ( -41.70, z-score = -3.76, R) >droEre2.scaffold_4820 3335382 108 - 10470090 GGCAUCGCAGCG--GCUAAGUAGCUUCAACACCGAAAACUAGUUUC--ACACACAGACUAGUUUGCACUCGAGCU--------CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC .((......))(--(.(((.((((.(((((((((.((((((((((.--......)))))))))).)...(((((.--------..))))))))))))).)))).))).)).......... ( -38.30, z-score = -4.42, R) >droSec1.super_33 176946 108 + 478481 GGCGGCGCAGCG--GCUAAGUAGCUUCAACACCGAAAACUAGUUUC--ACACACAGACUAGCUUGCACUCGAGCU--------CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC .((......))(--(.(((.((((.((((((((.....(((((((.--......)))))))........(((((.--------..))))))))))))).)))).))).)).......... ( -35.00, z-score = -2.53, R) >droSim1.chr3R 20745957 108 + 27517382 GGCGGCGCAGCG--GCUAAGUAGCUUCAACACCGAAAACUAGUUUC--ACACACAGACUAGCUUGCACUCGAGCU--------CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC .((......))(--(.(((.((((.((((((((.....(((((((.--......)))))))........(((((.--------..))))))))))))).)))).))).)).......... ( -35.00, z-score = -2.53, R) >droAna3.scaffold_13340 6778131 104 + 23697760 ------GCAUCA--GCUAAGUAGCUUCAACACCGAAAACUAGUUUCGCACACACAGACUAGUUUGCACUCGAGCU--------CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC ------((....--))(((.((((.(((((((((.((((((((((.(......))))))))))).)...(((((.--------..))))))))))))).)))).)))............. ( -33.60, z-score = -3.84, R) >consensus GGCGGCGCAGCG__GCUAAGUAGCUUCAACACCGAAAACUAGUUUC__ACACACAGACUAGUUUGCACUCGAGCU________CGGCUCGGGUGUUGAUGCUGAUUAACCAUAUUUAUAC ((.....((((...(((....))).(((((((((.((((((((((.........)))))))))).)...((((((.........)))))))))))))).)))).....)).......... (-34.30 = -35.13 + 0.83)

| Location | 20,915,810 – 20,915,918 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.86 |
| Shannon entropy | 0.12947 |
| G+C content | 0.48158 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -31.14 |
| Energy contribution | -30.92 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
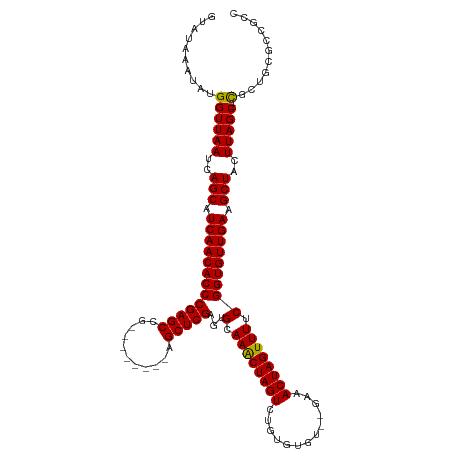
>dm3.chr3R 20915810 108 - 27905053 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG--------AGCUCGAGUGCAAACUAGUCUGUGUGU--GAAACUAGUUUUCGGUGUUGAAGCUACUUAGC--CGCUGCGCCGCC ..........((((((..(((.(((((((((((((..--------.)))))...(.((((((((........--...)))))))).))))))))).)))..)))))--)((......)). ( -36.30, z-score = -2.91, R) >droYak2.chr3R 3251328 118 + 28832112 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCGAGCACCCAAGCUCGAGUGCAAACUAGUCUGUGUGU--GAAACUAGUUUUCGGUGUUGAAGCUACUUAGCUUCGCUGCGAUGCC ..........(((....((((...((((((...(((((((......))))).))(.((((((((........--...)))))))).)))))))((((((....))))))))))....))) ( -38.50, z-score = -2.95, R) >droEre2.scaffold_4820 3335382 108 + 10470090 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG--------AGCUCGAGUGCAAACUAGUCUGUGUGU--GAAACUAGUUUUCGGUGUUGAAGCUACUUAGC--CGCUGCGAUGCC ((((.....(((((((..(((.(((((((((((((..--------.)))))...(.((((((((........--...)))))))).))))))))).)))..)))))--)).....)))). ( -38.10, z-score = -3.73, R) >droSec1.super_33 176946 108 - 478481 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG--------AGCUCGAGUGCAAGCUAGUCUGUGUGU--GAAACUAGUUUUCGGUGUUGAAGCUACUUAGC--CGCUGCGCCGCC ..........((((((..(((.(((((((((((((..--------.)))))...(.((((((((........--...)))))))).))))))))).)))..)))))--)((......)). ( -36.30, z-score = -2.48, R) >droSim1.chr3R 20745957 108 - 27517382 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG--------AGCUCGAGUGCAAGCUAGUCUGUGUGU--GAAACUAGUUUUCGGUGUUGAAGCUACUUAGC--CGCUGCGCCGCC ..........((((((..(((.(((((((((((((..--------.)))))...(.((((((((........--...)))))))).))))))))).)))..)))))--)((......)). ( -36.30, z-score = -2.48, R) >droAna3.scaffold_13340 6778131 104 - 23697760 GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG--------AGCUCGAGUGCAAACUAGUCUGUGUGUGCGAAACUAGUUUUCGGUGUUGAAGCUACUUAGC--UGAUGC------ ..........((((((..(((.(((((((((((((..--------.)))))...(.((((((((...((....))..)))))))).))))))))).)))..)))))--).....------ ( -35.10, z-score = -3.12, R) >consensus GUAUAAAUAUGGUUAAUCAGCAUCAACACCCGAGCCG________AGCUCGAGUGCAAACUAGUCUGUGUGU__GAAACUAGUUUUCGGUGUUGAAGCUACUUAGC__CGCUGCGCCGCC ...........(((((..(((.(((((((((((((...........)))))...(.((((((((.............)))))))).))))))))).)))..))))).............. (-31.14 = -30.92 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:28 2011