| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,906,314 – 20,906,407 |
| Length | 93 |
| Max. P | 0.647641 |

| Location | 20,906,314 – 20,906,407 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 58.70 |
| Shannon entropy | 0.72383 |
| G+C content | 0.52860 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -9.50 |
| Energy contribution | -10.45 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
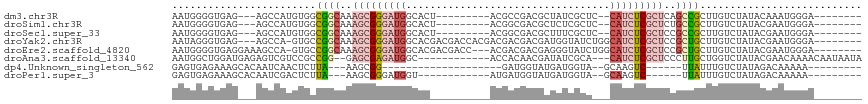
>dm3.chr3R 20906314 93 - 27905053 AAUGGGGUGAG---AGCCAUGUGGCGGCAAAGCGGGAUGGCACU---------ACGCCGACGCUAUCGCUC--CAUCUCGCUCAGCCGCUUGUCUAUACAAAUGGGA-------- .(((((((...---.)))....((((((..(((((((((((...---------..)))((.((....))))--.))))))))..))))))...))))..........-------- ( -35.70, z-score = -1.23, R) >droSim1.chr3R 20736456 93 - 27517382 AAUGGGGUGAG---AGCCAUGUGGCGGCAAAGCGGGAUGGCACU---------ACGGCGACGCUCUCGCUC--CAUCUCGCUCUGCCGCUUGUCUAUACGAAUGGGA-------- .(((((((...---.)))....(((((((.((((((((((....---------..(((((.....))))))--))))))))).)))))))...))))..........-------- ( -41.90, z-score = -2.73, R) >droSec1.super_33 167606 93 - 478481 AAUGGGGUGAG---AGCCAUGUGGCGGCAAAGCGGGAUGGCACU---------ACGGCGACGCUUUCGCUC--CAUCUCGCUCCGCCGCUUGUCUAUACGAAUGGGA-------- .(((((((...---.)))..(((((((...((((((((((....---------..(((((.....))))))--))))))))))))))))....))))..........-------- ( -40.90, z-score = -2.30, R) >droYak2.chr3R 3241065 103 + 28832112 AAUAGGGUGAG---AGCCA-GUGCCGGCAAAGCGGGAUGGCACGACGACCACGACGACGACGAUGGUAUCUGGCAUCUCGCUCCGCUGCUUGUCUAUACGAAUGGGA-------- .(((((((...---.))).-..((.(((..(((((((((.((.((..((((((.......)).)))).)))).)))))))))..)))))....))))..........-------- ( -35.30, z-score = -0.59, R) >droEre2.scaffold_4820 3323605 103 + 10470090 AAUGGGGUGAGGAAAGCCA-GUGCCGGCAAAGCGGGAUGGCACGACGACC---ACGACGACGAGGGUAUCUGGCAUCUCGCUCCGCUGCUUGUCUAUACGAAUGGGA-------- ...(((((((((...((((-(((((..(...(((((.((......)).))---.)).)...)..)))).))))).))))))))).((..((((....))))..))..-------- ( -34.40, z-score = -0.12, R) >droAna3.scaffold_13340 6769609 98 - 23697760 AAUGGCUGGAUGAGAGUCGUCCGCCGG--GAGCGAGAUGGC------------ACCACAACGAUAUCGCA---CAUCUCGCUCCCUUGCUGGUCUAUACGAACAAAACAAUAAUA ...(((.(((((.....))))))))((--((((((((((((------------..............)).---)))))))))))).............................. ( -38.04, z-score = -4.18, R) >dp4.Unknown_singleton_562 1744 75 - 2825 GAGUGAGAAAGCACAAUCAACUCUUA---AAGCGG--------------------GAUGGUAUGAUGGUA--GCAAGUC------UUAUUUGUCUAUAGACAAAAA--------- ((((((((..((((.(((((((.((.---......--------------------)).))).)))).)).--))...))------))))))(((....))).....--------- ( -14.90, z-score = -0.89, R) >droPer1.super_3 6982609 83 + 7375914 GAGUGAGAAAGCACAAUCGACUCUUA---AAGCGGGAUGGU------------AUGAUGGUAUGAUGGUA--GCAAGUC------UUAUUUGUCUAUAGACAAAAA--------- ((((((((..((((.(((((((.(((---.(.(.....).)------------.))).))).)))).)).--))...))------))))))(((....))).....--------- ( -15.70, z-score = -0.27, R) >consensus AAUGGGGUGAG___AGCCAUGUGCCGGCAAAGCGGGAUGGCAC__________ACGACGACGAUAUCGCA___CAUCUCGCUCCGCUGCUUGUCUAUACGAAUGGGA________ ........................((((..(((((((((..................................)))))))))..))))........................... ( -9.50 = -10.45 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:26 2011