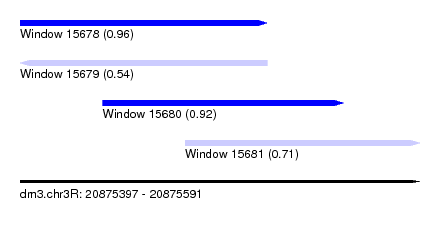
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,875,397 – 20,875,591 |
| Length | 194 |
| Max. P | 0.959768 |

| Location | 20,875,397 – 20,875,517 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Shannon entropy | 0.03817 |
| G+C content | 0.44333 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
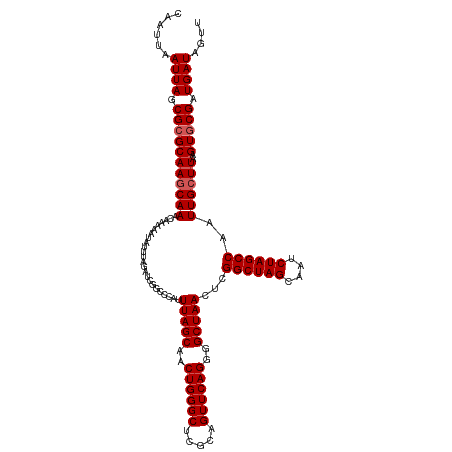
>dm3.chr3R 20875397 120 + 27905053 CAAUUAAUUAGCGGGCAAGCAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCUCGGAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUU ......((((.((.((((((((........................(((((..((((((.....))))))..)))))...((((((....))))))..))))))..)).)).)))).... ( -34.20, z-score = -1.41, R) >droSim1.chr3R 20702787 120 + 27517382 CAAUUAAUUAGCGCGCAAACAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUU ......((((.(((((.......................((((((.........)))))).((((((..(((.....)))((((((....))))))))))))....))))).)))).... ( -34.40, z-score = -1.59, R) >droSec1.super_33 137157 120 + 478481 CAAUUAAUUAGCGCGCAAGCAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUU ......((((.(((((((((((........................(((((..((((((.....))))))..)))))...((((((....))))))..))))))..))))).)))).... ( -38.00, z-score = -2.17, R) >droYak2.chr3R 3209697 120 - 28832112 CAAUUAAUUAGCGCGCAAGCAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUU ......((((.(((((((((((........................(((((..((((((.....))))))..)))))...((((((....))))))..))))))..))))).)))).... ( -38.00, z-score = -2.49, R) >droEre2.scaffold_4820 3292783 120 - 10470090 CAAUUAAUUAGCGCGCAAGCAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCCCACAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUU ......((((.(((((((((((........................(((((..((((((.....))))))..)))))...((((((....))))))..))))))..))))).)))).... ( -37.30, z-score = -2.59, R) >consensus CAAUUAAUUAGCGCGCAAGCAAACAAAAAUAUUUAGAUCGGCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUU ......((((.(((((((((((........................(((((..((((((.....))))))..)))))...((((((....))))))..))))))..))))).)))).... (-35.40 = -35.80 + 0.40)

| Location | 20,875,397 – 20,875,517 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Shannon entropy | 0.03817 |
| G+C content | 0.44333 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20875397 120 - 27905053 AACUAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUCCGAGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGCUUGCCCGCUAAUUAAUUG ..........((...(((((((..((((((....))))))(((((........)))))((.(((((.........)))))))................)))))))...)).......... ( -31.30, z-score = -1.85, R) >droSim1.chr3R 20702787 120 - 27517382 AACCAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUGCGAGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGUUUGCGCGCUAAUUAAUUG .........((((..(((((((((((((((....))))))))((..((((..((((((......))))).).....))))..))..........)))))))..))))............. ( -32.30, z-score = -1.34, R) >droSec1.super_33 137157 120 - 478481 AACCAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUGCGAGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGCUUGCGCGCUAAUUAAUUG ..........((.(.(((((((((((((((....))))))))((..((((..((((((......))))).).....))))..))..............))))))).).)).......... ( -31.90, z-score = -0.97, R) >droYak2.chr3R 3209697 120 + 28832112 AACUAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUGCGAGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGCUUGCGCGCUAAUUAAUUG ..........((.(.(((((((((((((((....))))))))((..((((..((((((......))))).).....))))..))..............))))))).).)).......... ( -31.90, z-score = -0.95, R) >droEre2.scaffold_4820 3292783 120 + 10470090 AACUAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUGUGGGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGCUUGCGCGCUAAUUAAUUG ..........((.(.(((((((((((((((....))))))))...............((.((((((.........)))))).))..............))))))).).)).......... ( -33.80, z-score = -1.14, R) >consensus AACUAUCAUCGCACUUAAGCAAUUGGCUAGAUUGCUAGCCGAGUUAGCCCCUGAACUGCGAGCCCAGUUGCUAAAUGGGCCGAUCUAAAUAUUUUUGUUUGCUUGCGCGCUAAUUAAUUG ..........((.(.(((((((((((((((....))))))))................((.(((((.........)))))))................))))))).).)).......... (-30.38 = -30.26 + -0.12)

| Location | 20,875,437 – 20,875,554 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 96.92 |
| Shannon entropy | 0.05362 |
| G+C content | 0.48718 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -38.58 |
| Energy contribution | -39.38 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
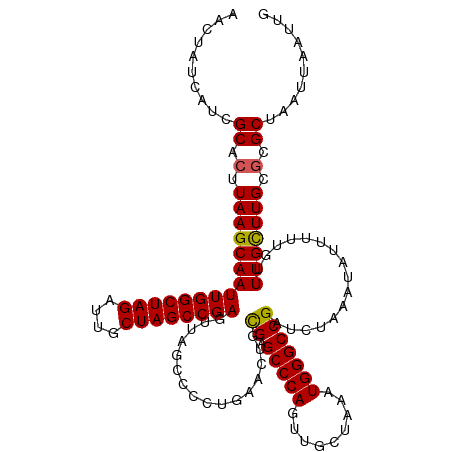
>dm3.chr3R 20875437 117 + 27905053 GCCCAUUUAGCAACUGGGCUCGGAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUU (((...(((((..((((((.....))))))..))))).((((((((....)))))).(((((......)))))))......)))..(((((((((........).)))))))).... ( -42.00, z-score = -2.34, R) >droSim1.chr3R 20702827 117 + 27517382 GCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUU (((...(((((..((((((.....))))))..))))).((((((((....)))))).(((((......)))))))......)))..(((((((((........).)))))))).... ( -42.00, z-score = -2.01, R) >droSec1.super_33 137197 117 + 478481 GCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUU (((...(((((..((((((.....))))))..))))).((((((((....)))))).(((((......)))))))......)))..(((((((((........).)))))))).... ( -42.00, z-score = -2.01, R) >droYak2.chr3R 3209737 117 - 28832112 GCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGUACGCUCAAAUUAAAGCAGCGUUUCGUUU (((...(((((..((((((.....))))))..))))).((((((((....)))))).(((((......)))))))......)))..((.((((((........).))))).)).... ( -38.10, z-score = -1.15, R) >droEre2.scaffold_4820 3292823 117 - 10470090 GCCCAUUUAGCAACUGGGCCCACAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUAACGUGGUACGCUCAAAUUAAAGCAGCGUUUCGUAU ......(((((..((((((.....))))))..)))))...((((((....)))))).(((((......))))).........(((....((((((........).)))))..))).. ( -34.40, z-score = -1.05, R) >consensus GCCCAUUUAGCAACUGGGCUCGCAGUUCAGGGGCUAACUCGGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUU (((...(((((..((((((.....))))))..))))).((((((((....)))))).(((((......)))))))......)))..(((((((((........).)))))))).... (-38.58 = -39.38 + 0.80)

| Location | 20,875,477 – 20,875,591 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.78 |
| Shannon entropy | 0.19322 |
| G+C content | 0.46646 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -28.58 |
| Energy contribution | -30.50 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20875477 114 + 27905053 GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUUCGUAAAGCGUGAGAUUCGCGAUCGUGUUGUUAGGCAG--- ((((((....))))))...((((((((..((((..((((((((((.....))).))))))).....(((..((....((.....))...))..))).))))..)..))))))).--- ( -38.90, z-score = -2.18, R) >droSim1.chr3R 20702867 117 + 27517382 GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUUCGUAAAGCGUGAGAUUCGCGAUCGUGUUGUUAGGCAGUUU ((((((....))))))(((((((((((..((((....((((.(((((((((((((........).)))))))))...))).))))(((.....))).))))..)..)))))))))). ( -40.90, z-score = -2.47, R) >droSec1.super_33 137237 114 + 478481 GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUGGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUUCGUAAAGCGUGAGAUUCGCGAUCGUGUUGUUAGGCAG--- ((((((....))))))...((((((((..((((....((((.(((((((((((((........).)))))))))...))).))))(((.....))).))))..)..))))))).--- ( -38.60, z-score = -1.87, R) >droYak2.chr3R 3209777 112 - 28832112 GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGUACGCUCAAAUUAAAGCAGCGUUUCGUUUCGUAAGGCGUGAGAUUCGCGAUCGUGUUUGGGGAG----- ((((((....)))))).....((((((..((((..((((((((((.....))).))))))).....((((((((..((....))..)))))..))).))))..))))))...----- ( -39.40, z-score = -2.46, R) >droEre2.scaffold_4820 3292863 100 - 10470090 GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUAACGUGGUACGCUCAAAUUAAAGCAGCGUUUCGUAUCGU------------UCGCGAUCGUGUUUUUGGGG----- ((((((....)))))).....((((((.((.(((((.((..(((((((.((((((........).))))).)))...)))------------).)).))))))).)))))).----- ( -26.10, z-score = -0.36, R) >consensus GGCUAGCAAUCUAGCCAAUUGCUUAAGUGCGAUGAUAGUUUGGCGUGGAACGCUCAAAUUAAAGCAGCGUUUCGUUUCGUAAAGCGUGAGAUUCGCGAUCGUGUUGUUAGGCAG___ ((((((....))))))...((((((((..((((..((((((((((.....))).))))))).....(((..((....((.....))...))..))).))))..))..)))))).... (-28.58 = -30.50 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:22 2011