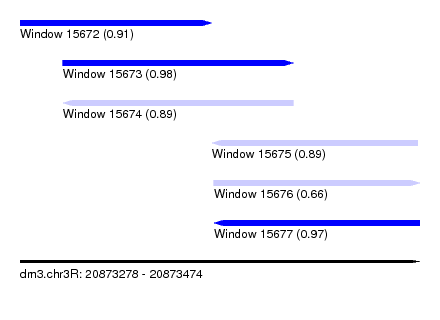
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,873,278 – 20,873,474 |
| Length | 196 |
| Max. P | 0.975014 |

| Location | 20,873,278 – 20,873,372 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.10 |
| Shannon entropy | 0.48452 |
| G+C content | 0.40931 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -13.79 |
| Energy contribution | -15.23 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
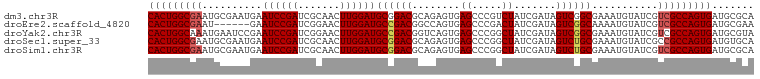
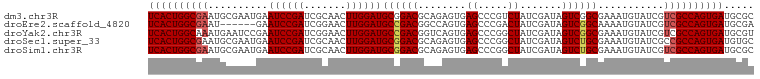
>dm3.chr3R 20873278 94 + 27905053 -------------------CAGUACUCAUUAAGUAG-----UCGGCCUAUCUACCAGAAUAUUAGCCAAGGUACACAUUUGUACCUGCAAACGUACCUAGGUACGAAAGCUGUCACCU -------------------...((((.....))))(-----.((((..................((..(((((((....)))))))))...(((((....)))))...)))).).... ( -23.40, z-score = -1.83, R) >droEre2.scaffold_4820 3290680 117 - 10470090 CUGUGCUUAUCAGGCAGUACAAGAUCAGUUAGACAAAAGUUUUUCCUUUGCCACCAAA-UAUCAACCUAGGUACAUGUGUGUACCUGCUACCUUACCUAGGUACGAAAGCUGUCACCU .((((((........)))))).((.(((((.(.((((.(.....).)))).)......-.....(((((((((...(((.((....)))))..))))))))).....))))))).... ( -25.10, z-score = -0.07, R) >droSim1.chr3R 20700635 118 + 27517382 CAGUACUAAUUAAGCAGUUCAAGUCGAGUUAAACAAAAGUUUAUUCCUAUUUACCAAAAUAUUAGCCUAGGUACAGAUUUGUACCUGCAAACGUACCUAGGUUCGAAAGCUGUCACCU .............((((((....((((..(((((....))))).....................((((((((((.(.(((((....)))))))))))))))))))).))))))..... ( -29.20, z-score = -3.49, R) >consensus C_GU_CU_AU_A_GCAGU_CAAGACCAGUUAAACAAAAGUUUAUCCCUAUCUACCAAAAUAUUAGCCUAGGUACACAUUUGUACCUGCAAACGUACCUAGGUACGAAAGCUGUCACCU ................................................................((((((((((...(((((....))))).)))))))))).(....)......... (-13.79 = -15.23 + 1.45)

| Location | 20,873,299 – 20,873,412 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Shannon entropy | 0.30320 |
| G+C content | 0.44173 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -18.07 |
| Energy contribution | -17.87 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20873299 113 + 27905053 -----CCUAUCUACCAGAAUAUUAGCCAAGGUACACAUUUGUACCUGCAAACGUACCUAGGUACGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAU -----...........((......((((((((((((.((((((((((..........)))))))))).).))).)))))).)).....((((((.........))))))....))... ( -31.30, z-score = -3.06, R) >droEre2.scaffold_4820 3290720 117 - 10470090 UUUUCCUUUGCCACCAAA-UAUCAACCUAGGUACAUGUGUGUACCUGCUACCUUACCUAGGUACGAAAGCUGUCACCUUUCGCAUCACUGGCGACGAUACAUUUUGCCGACUAUCGAU .......((((((.....-.....(((((((((...(((.((....)))))..))))))))).((((((.......))))))......))))))(((((............))))).. ( -28.10, z-score = -2.38, R) >droYak2.chr3R 3207589 115 - 28832112 --UUUCUUUCCCACCAAAAUAUCAUCAUAGGUACACACUUGUACCUAC-GCCUUACCUAGGUACGAAAGCUGUCACCUUACGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAU --...(((((.................((((((((....)))))))).-((((.....))))..)))))..(((......(((.......))).(((......)))..)))....... ( -23.80, z-score = -2.47, R) >droSec1.super_33 135055 112 + 478481 UUAUUCCUAUUUACCAAAAUAUUACCCUAGGUACAGAUUUGUACCUG------UAUCUAGGUACGAAAGCUGCAACCUUGCACAUCACUGGCGGCGAUACAUUUCGCAGACUAUCGAU .........................(((((((((((........)))------))))))))..(((.((((((....((((.((....)))))).((......)))))).)).))).. ( -29.40, z-score = -3.16, R) >droSim1.chr3R 20700675 118 + 27517382 UUAUUCCUAUUUACCAAAAUAUUAGCCUAGGUACAGAUUUGUACCUGCAAACGUACCUAGGUUCGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCAGACUAUCGAU ........................((((((((((.(.(((((....))))))))))))))))((((.((((((.......(((.......)))..((......)))))).)).)))). ( -30.70, z-score = -2.78, R) >consensus UUAUUCCUAUCUACCAAAAUAUUAGCCUAGGUACACAUUUGUACCUGC_AACGUACCUAGGUACGAAAGCUGUCACCUUGCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAU ............(((............((((((((....))))))))............))).(((.(((.(((.((............)).)))((......))...).)).))).. (-18.07 = -17.87 + -0.20)

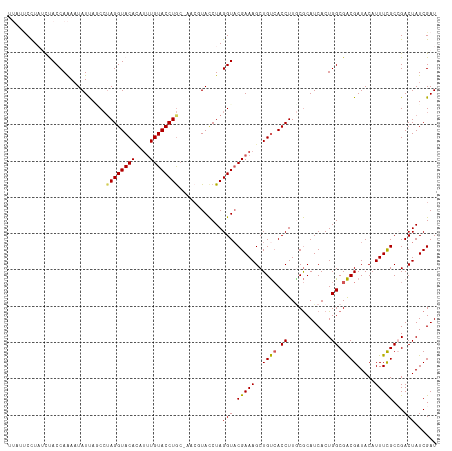
| Location | 20,873,299 – 20,873,412 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Shannon entropy | 0.30320 |
| G+C content | 0.44173 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20873299 113 - 27905053 AUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGUACCUAGGUACGUUUGCAGGUACAAAUGUGUACCUUGGCUAAUAUUCUGGUAGAUAGG----- .......((((((((......)))))(((((....(((((((....)......(((((....))))).))))(((((((....)))))))..)).......))))).)))...----- ( -35.50, z-score = -1.55, R) >droEre2.scaffold_4820 3290720 117 + 10470090 AUCGAUAGUCGGCAAAAUGUAUCGUCGCCAGUGAUGCGAAAGGUGACAGCUUUCGUACCUAGGUAAGGUAGCAGGUACACACAUGUACCUAGGUUGAUA-UUUGGUGGCAAAGGAAAA .......((((.(((...(((((((((((............)))))).(((.((.(((....))).)).)))(((((((....))))))).....))))-)))).))))......... ( -33.70, z-score = -1.55, R) >droYak2.chr3R 3207589 115 + 28832112 AUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGUAAGGUGACAGCUUUCGUACCUAGGUAAGGC-GUAGGUACAAGUGUGUACCUAUGAUGAUAUUUUGGUGGGAAAGAAA-- .......((((.(...((((((((.(....)))))))))..).))))..(((((.((((.(((((...(-(((((((((....))))))))))....))))).)))).)))))...-- ( -41.00, z-score = -4.02, R) >droSec1.super_33 135055 112 - 478481 AUCGAUAGUCUGCGAAAUGUAUCGCCGCCAGUGAUGUGCAAGGUUGCAGCUUUCGUACCUAGAUA------CAGGUACAAAUCUGUACCUAGGGUAAUAUUUUGGUAAAUAGGAAUAA ..(((.((.((((((..((((((((.....)))))..)))...)))))))).)))..(((((.((------(((((....))))))).)))))....((((((........)))))). ( -32.90, z-score = -2.19, R) >droSim1.chr3R 20700675 118 - 27517382 AUCGAUAGUCUGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGAACCUAGGUACGUUUGCAGGUACAAAUCUGUACCUAGGCUAAUAUUUUGGUAAAUAGGAAUAA .....((((((((.(((((((((...((((....)).)).((((............)))).)))))))))))(((((((....))))))))))))).((((((........)))))). ( -33.90, z-score = -1.70, R) >consensus AUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCAAGGUGACAGCUUUCGUACCUAGGUAAGGU_GCAGGUACAAAUCUGUACCUAGGCUAAUAUUUUGGUAGAUAGGAAAAA ..(((.(((..((.....))...((((((............)))))).))).)))((((.(((((.......(((((((....))))))).......))))).))))........... (-23.38 = -23.58 + 0.20)

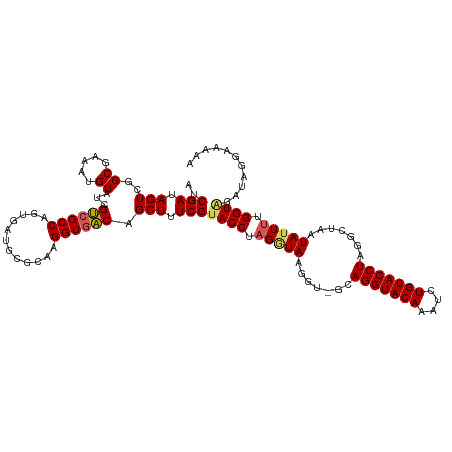
| Location | 20,873,372 – 20,873,473 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Shannon entropy | 0.19240 |
| G+C content | 0.55106 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -29.18 |
| Energy contribution | -29.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20873372 101 - 27905053 CACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGUCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCA (((((((((.(((((.((....)).))))).....((((((..(((...))).(((.(.(((....))).))))....)))))).)))))))))....... ( -35.40, z-score = -0.70, R) >droEre2.scaffold_4820 3290797 95 + 10470090 CACUGGCGAAU------GAAUCCGAUCGGAACUUGGAUGCCGACGGCCAGUGAGCCCGACUAUCGAUAGUCGGCAAAAUGUAUCGUCGCCAGUGAUGCGAA (((((((((.(------((((((((.......))))))(((((((((......)))((.....))...))))))........))))))))))))....... ( -39.30, z-score = -3.60, R) >droYak2.chr3R 3207664 101 + 28832112 CACUGGCAAAUGAAUCCGAAUCCGAUCGGAACUUGGAUGCCGACGGUCAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGUA (((((((......((((((.(((....)))..))))))...((((((((....(((.(((((....))))))))....)).)))))))))))))....... ( -41.30, z-score = -3.01, R) >droSec1.super_33 135127 101 - 478481 CACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGCCGCCAGUGAUGUGCA ((((((((..(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...))))))..))))))))....... ( -38.80, z-score = -2.17, R) >droSim1.chr3R 20700753 101 - 27517382 CACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGUCGCCAGUGAUGCGCA (((((((((.(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...)))))).)))))))))....... ( -40.30, z-score = -2.30, R) >consensus CACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGCA (((((((((..........((((((.......))))))((((((........((.....)).......))))))...........)))))))))....... (-29.18 = -29.46 + 0.28)
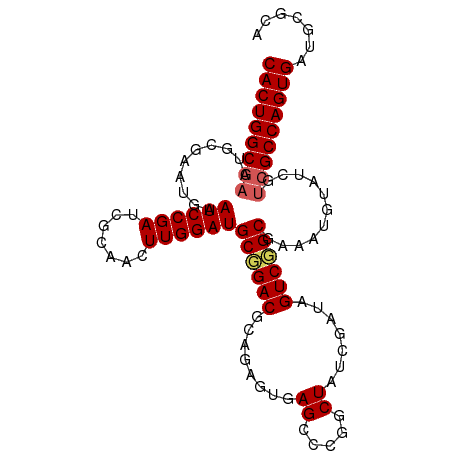
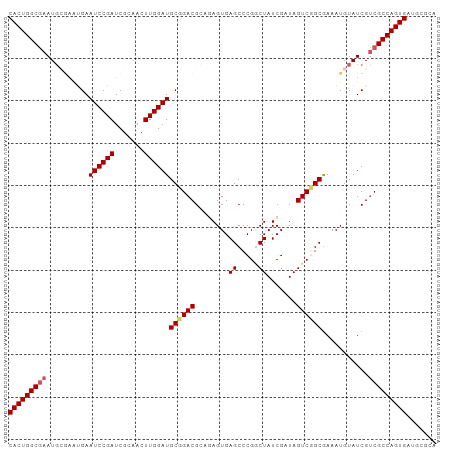
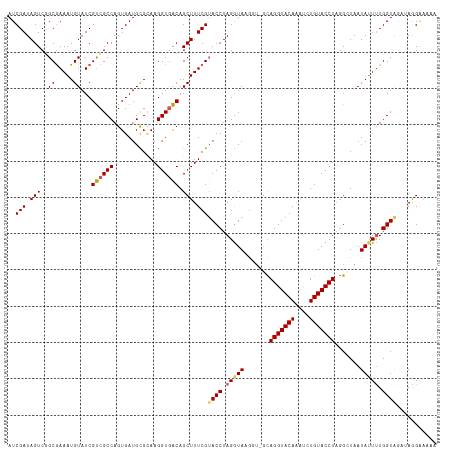
| Location | 20,873,373 – 20,873,474 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.50 |
| Shannon entropy | 0.19240 |
| G+C content | 0.55106 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20873373 101 + 27905053 GCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGACGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGA .....(((((((((((((......((((.((((...(((.(.(((((.(.....).)))))))))..)))))))))))((.....))....)))))))))) ( -34.30, z-score = -1.45, R) >droEre2.scaffold_4820 3290798 95 - 10470090 UCGCAUCACUGGCGACGAUACAUUUUGCCGACUAUCGAUAGUCGGGCUCACUGGCCGUCGGCAUCCAAGUUCCGAUCGGAUUC------AUUCGCCAGUGA .....((((((((((..........(((((((...((.....))((((....))))))))))).......(((....)))...------..)))))))))) ( -38.40, z-score = -3.36, R) >droYak2.chr3R 3207665 101 - 28832112 ACGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGCCGGGCUCACUGACCGUCGGCAUCCAAGUUCCGAUCGGAUUCGGAUUCAUUUGCCAGUGA .....((((((((((...........((((((..(((..(((...)))...)))..))))))........(((((......))))).....)))))))))) ( -36.00, z-score = -2.19, R) >droSec1.super_33 135128 101 + 478481 GCACAUCACUGGCGGCGAUACAUUUCGCAGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGA .....(((((((((((((......)))).((((...(((.((.((((.(.....).)))))))))..))))((((..........))))...))))))))) ( -36.70, z-score = -2.43, R) >droSim1.chr3R 20700754 101 + 27517382 GCGCAUCACUGGCGACGAUACAUUUCGCAGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGA .....(((((((((((((......((((((.((...(((.((.((((.(.....).)))))))))..)))))))))))((.....))....)))))))))) ( -34.60, z-score = -1.57, R) >consensus GCGCAUCACUGGCGACGAUACAUUUCGCCGACUAUCGAUAGCCGGGCUCACUCUGCGUCCGCAUCCAAGUUGCGAUCGGAUUCAUUCGCAUUCGCCAGUGA .....(((((((((((((......)))..((((...(((.(.(((((.........)))))))))..))))....................)))))))))) (-24.88 = -24.80 + -0.08)
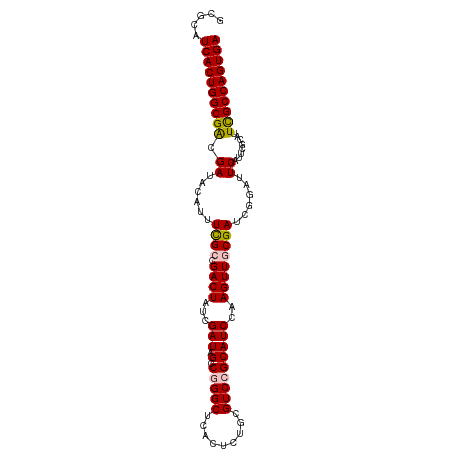
| Location | 20,873,373 – 20,873,474 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Shannon entropy | 0.19240 |
| G+C content | 0.55106 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -30.58 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
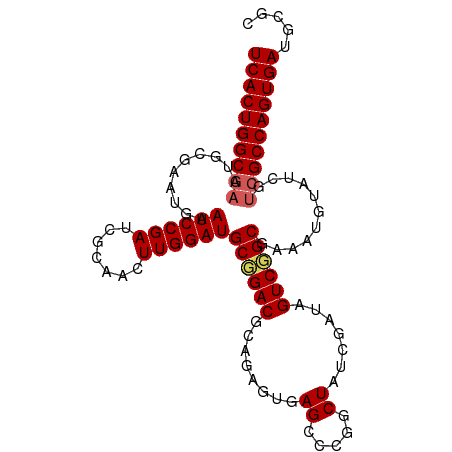
>dm3.chr3R 20873373 101 - 27905053 UCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGUCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGC ((((((((((.(((((.((....)).))))).....((((((..(((...))).(((.(.(((....))).))))....)))))).))))))))))..... ( -36.80, z-score = -1.19, R) >droEre2.scaffold_4820 3290798 95 + 10470090 UCACUGGCGAAU------GAAUCCGAUCGGAACUUGGAUGCCGACGGCCAGUGAGCCCGACUAUCGAUAGUCGGCAAAAUGUAUCGUCGCCAGUGAUGCGA ((((((((((.(------((((((((.......))))))(((((((((......)))((.....))...))))))........)))))))))))))..... ( -40.70, z-score = -3.77, R) >droYak2.chr3R 3207665 101 + 28832112 UCACUGGCAAAUGAAUCCGAAUCCGAUCGGAACUUGGAUGCCGACGGUCAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGU ((((((((......((((((.(((....)))..))))))...((((((((....(((.(((((....))))))))....)).))))))))))))))..... ( -42.70, z-score = -3.29, R) >droSec1.super_33 135128 101 - 478481 UCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGCCGCCAGUGAUGUGC (((((((((..(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...))))))..)))))))))..... ( -40.20, z-score = -2.71, R) >droSim1.chr3R 20700754 101 - 27517382 UCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCUGCGAAAUGUAUCGUCGCCAGUGAUGCGC ((((((((((.(((((.((....)).))))).....((((((..((((((.(((((...))).....)).))))))...)))))).))))))))))..... ( -41.70, z-score = -2.82, R) >consensus UCACUGGCGAAUGCGAAUGAAUCCGAUCGCAACUUGGAUGCGGACGCAGAGUGAGCCCGGCUAUCGAUAGUCGGCGAAAUGUAUCGUCGCCAGUGAUGCGC ((((((((((..........((((((.......))))))((((((........((.....)).......))))))...........))))))))))..... (-30.58 = -30.86 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:19 2011