| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,870,545 – 20,870,619 |
| Length | 74 |
| Max. P | 0.950747 |

| Location | 20,870,545 – 20,870,619 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Shannon entropy | 0.27691 |
| G+C content | 0.39421 |
| Mean single sequence MFE | -19.35 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.15 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.594347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
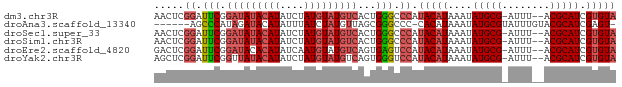
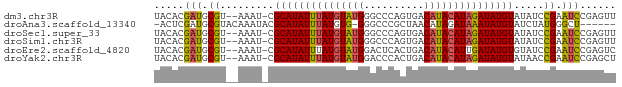
>dm3.chr3R 20870545 74 + 27905053 AACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCG-AUUU--ACGCAUCGUGUA ..(((((.....(((((((((....))))))))).)))))....(((((....(((((-....--.))))).))))) ( -19.20, z-score = -1.65, R) >droAna3.scaffold_13340 6229584 69 - 23697760 ------AGCCCAUAGAUACAUAUUUAUCUAUGUUAGCGGGCCC-CACAUAAAUAUGCGUAUUUGUACGCAUCGAGU- ------.(((((((((((......)))))))(....)))))..-.........(((((((....))))))).....- ( -18.60, z-score = -2.47, R) >droSec1.super_33 132284 74 + 478481 AACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCG-AUUU--ACGCAUCGUGUA ..(((((.....(((((((((....))))))))).)))))....(((((....(((((-....--.))))).))))) ( -19.20, z-score = -1.65, R) >droSim1.chr3R 20697922 74 + 27517382 AACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCG-AUUU--ACGCAUCGUGUA ..(((((.....(((((((((....))))))))).)))))....(((((....(((((-....--.))))).))))) ( -19.20, z-score = -1.65, R) >droEre2.scaffold_4820 3287885 74 - 10470090 GACUCGGAUUCGGAUACACAUAUCAAUGUAUGUCAGUGAGUCCAUACAUAAAUAUGCG-AUUU--ACGCAUCGUGUA .....((((((.((((.((((....)))).))))...)))))).(((((....(((((-....--.))))).))))) ( -20.50, z-score = -1.83, R) >droYak2.chr3R 3204938 74 - 28832112 AGCUCGGAUUCGGUUAUACAUAUCUAUGUAUGUCAGUGGGUCCAUACAUAAAUAUGCG-AUUU--ACGCAUCGUGUA .....(((((((.((((((((....)))))))..).))))))).(((((....(((((-....--.))))).))))) ( -19.40, z-score = -1.44, R) >consensus AACUCGGAUUCGGAUAUACAUAUCUAUGUAUGUCACUGGGCCCAUACAUAAAUAUGCG_AUUU__ACGCAUCGUGUA .....((.(((.(((((((((....)))))))))...))).)).(((((....(((((........))))).))))) (-13.06 = -14.15 + 1.09)
| Location | 20,870,545 – 20,870,619 |
|---|---|
| Length | 74 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Shannon entropy | 0.27691 |
| G+C content | 0.39421 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
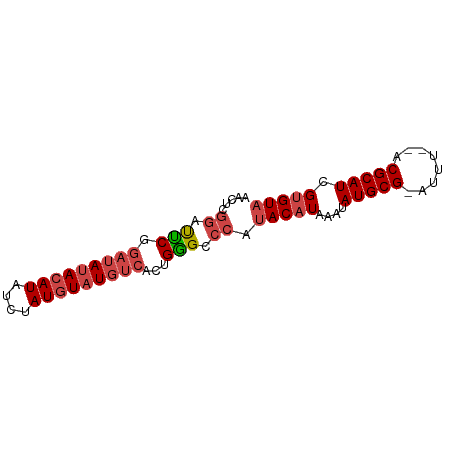
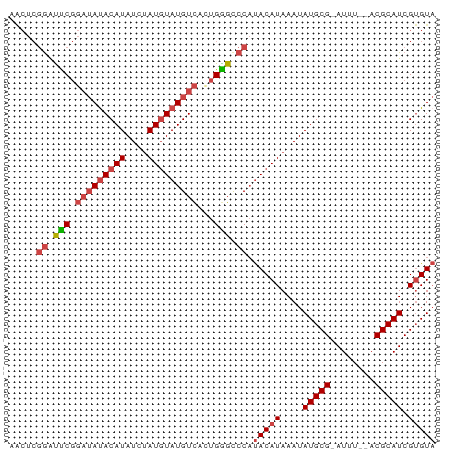
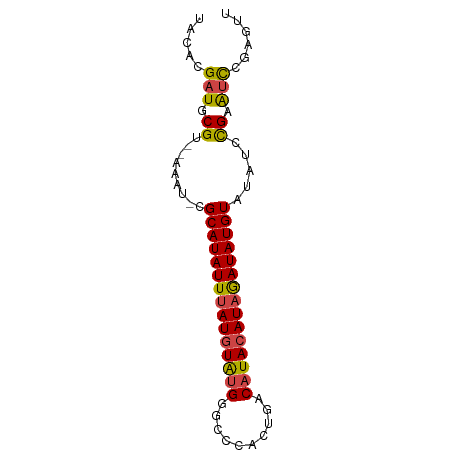
>dm3.chr3R 20870545 74 - 27905053 UACACGAUGCGU--AAAU-CGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUU .....(((.((.--....-.(((((((((((((((..........))))))))))))))).....)).)))...... ( -19.20, z-score = -1.81, R) >droAna3.scaffold_13340 6229584 69 + 23697760 -ACUCGAUGCGUACAAAUACGCAUAUUUAUGUG-GGGCCCGCUAACAUAGAUAAAUAUGUAUCUAUGGGCU------ -.....(((((((....))))))).........-.(((((......(((((((......))))))))))))------ ( -18.90, z-score = -1.79, R) >droSec1.super_33 132284 74 - 478481 UACACGAUGCGU--AAAU-CGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUU .....(((.((.--....-.(((((((((((((((..........))))))))))))))).....)).)))...... ( -19.20, z-score = -1.81, R) >droSim1.chr3R 20697922 74 - 27517382 UACACGAUGCGU--AAAU-CGCAUAUUUAUGUAUGGGCCCAGUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUU .....(((.((.--....-.(((((((((((((((..........))))))))))))))).....)).)))...... ( -19.20, z-score = -1.81, R) >droEre2.scaffold_4820 3287885 74 + 10470090 UACACGAUGCGU--AAAU-CGCAUAUUUAUGUAUGGACUCACUGACAUACAUUGAUAUGUGUAUCCGAAUCCGAGUC .....(((.((.--....-((((((((.(((((((..........))))))).))))))))....)).)))...... ( -18.60, z-score = -1.37, R) >droYak2.chr3R 3204938 74 + 28832112 UACACGAUGCGU--AAAU-CGCAUAUUUAUGUAUGGACCCACUGACAUACAUAGAUAUGUAUAACCGAAUCCGAGCU .....(((.((.--....-.(((((((((((((((..........))))))))))))))).....)).)))...... ( -19.20, z-score = -3.32, R) >consensus UACACGAUGCGU__AAAU_CGCAUAUUUAUGUAUGGGCCCACUGACAUACAUAGAUAUGUAUAUCCGAAUCCGAGUU ....................(((((((((((((((..........)))))))))))))))................. (-15.74 = -15.97 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:15 2011