
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,862,791 – 20,862,910 |
| Length | 119 |
| Max. P | 0.573470 |

| Location | 20,862,791 – 20,862,910 |
|---|---|
| Length | 119 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.29 |
| Shannon entropy | 0.74533 |
| G+C content | 0.48259 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -10.26 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20862791 119 - 27905053 GUGAUAGCAG-GCACACAUUGACGCCUUCUAUCUGGCCAUCGAUGAUCACAUCGGAUGGUUUUCCAAACGGCGUGCAUACUGCAUACUCCAUGCGCUCUGCCAAGUAGAUGGGCUUGUCC ......((((-((...((((.(((((........(((((((((((....)))).)))))))........)))))(((.(.(((((.....))))).).)))......)))).)))))).. ( -36.59, z-score = -0.63, R) >droSim1.chr3R 20690621 119 - 27517382 GCGAUAGCAG-GCACACAUUGACGCCUUCUAUCUGGCCAUCGAUGAUCACAUCGGAUGGUUUUCCAAACGGCGUGCAUACUGCAUACUCCAUGCGCUCCGCCAAGUAGAUGGGCUUGUCC ..(((((.((-((..........)))).))))).(.(((((((((....))))((.(((....)))..(((.((((((............)))))).))))).....))))).)...... ( -38.30, z-score = -1.09, R) >droSec1.super_33 124556 119 - 478481 GCGAUAGCAG-GCACACAUUGACGCCUUCUAUCUGGCCAUCGAUGAUCACAUCGGAUGGUUUUCCAAACGGCGUGCAUACUGCAUACUCCAUGCGCUCCGCCAAGUAGAUGGGCUUGUCC ..(((((.((-((..........)))).))))).(.(((((((((....))))((.(((....)))..(((.((((((............)))))).))))).....))))).)...... ( -38.30, z-score = -1.09, R) >droYak2.chr3R 3197149 119 + 28832112 GCGACAGCAG-ACACACAUUGACGCCUUCUAUCUGGCCAUCGAUGAGCACAUCGGAUGGUUUUCCAAACGGCGUGCAUACUGCAUACUCCAUGCGCUCCGCCAAAUAGAUGGGCUUGUCC ..((((((((-.....((((((.(((........)))..)))))).((((..((..(((....)))..))..))))...))))...........((.(((.(.....).))))).)))). ( -37.90, z-score = -1.57, R) >droEre2.scaffold_4820 3279950 119 + 10470090 GCGACAGCAG-ACACACAUUUACGCCUUCUAUCUGGCCAUCGAUGAGCACAUCGGAUGGUUUUCCAAACGGCGUGCAUACAGCAUACUCCAUGCGCUCCGCCAAAUAGAUGGGCUUGUCC ((....))..-............(((((((((.((((..((((((....)))))).(((....)))...((.((((((..((....))..)))))).)))))).))))).))))...... ( -37.00, z-score = -1.81, R) >droAna3.scaffold_13340 6222001 119 + 23697760 UGGACAGGAG-GCAUACUCUAACGCCAUCGAUCUCGCCGUCGAUAAGCACAUCUGAUGGUUUCCCGAAAGGUGUGCAGACAGCAUAUUCCAUGCGGUCCUUUAGCAACAGCGGAGAGUCC .((((.((((-.....))))..(((.((((((......))))))..((((((((..(((....)))..)))))))).(((.((((.....)))).)))...........)))....)))) ( -39.20, z-score = -1.66, R) >dp4.chr2 12585115 119 + 30794189 UACAGAGAAGGGCACAUU-UGACGCCAUCGAUUAAGCCUUCAAUUAUAAUGUCAGAAGGUUUUCCAAAAGGCGUACAUACAGCAUAUUCCAUUCGAUCUUGUAGUGAGAUUGUUGUAUCC (((((.(((..((.....-(((((((.......((((((((.............)))))))).......))))).))....))...)))....(((((((.....))))))))))))... ( -28.16, z-score = -1.82, R) >droPer1.super_3 6815467 119 + 7375914 UACAGAGAAGGGCACAUU-UUACGCCAUCGAUUAAGCCUUCAAUUAUAAUGUCAGAAGGUUUUCCAAAAGGCGUACAUACAGCAUAUUCCAUUCGAUCUUGUAGUGAGAUUGUUGUAUCC (((((.(((..((.....-.((((((.......((((((((.............)))))))).......))))))......))...)))....(((((((.....))))))))))))... ( -28.26, z-score = -2.34, R) >droWil1.scaffold_181130 13849906 119 - 16660200 GCGAUAGGAGAGCACACU-UGACACCUUCUAUCUGACCUUCAAUUAAAAUAUCGGACGGUUUUCCAAAUGGGGUGCACACAGCGUAUUCCAUUCGAUCCGUAAGCACAAUUGGUGUAUCG ..((((((((..((....-))....))))))))..((..((((((.......(((((((....)).((((((((((.......)))))))))).).)))).......)))))).)).... ( -27.34, z-score = -0.34, R) >droVir3.scaffold_12936 510543 119 - 632223 GCGAUAGAAGGGCACAAU-UGACAUCUUCAAUUUUUCCCUCAAUUAUAAUAUCUGAAGGUUUUCCGAAUGGCGUACAUACAGCGCAUUCCCUUCUAUCGCUUAGUACAAUAGGUGUAUCG ((((((((((((..((.(-((..(((((((.......................)))))))....))).))((((.......))))...))))))))))))...(((((.....))))).. ( -32.70, z-score = -3.63, R) >droMoj3.scaffold_6540 4240015 119 - 34148556 GCGAUAGCAGCGCACAGU-UGCAGUUUUCAAUUUUCCCUUCAAUGAUAAUAUCGGAAGGUUUUCCGAAUGGUGUGCAGACAGCACAUUCCUGCCUAUCCUUUAGCACAAUUGGUGUAUCG .((((((((((.....))-)))..............(((((...(((...))).)))))....((((.((((((((.....))))))...(((.((.....))))))).))))..))))) ( -27.40, z-score = -0.29, R) >droGri2.scaffold_15116 1329572 119 + 1808639 GCGAUAGAAGAGCACAGU-UCACAUUUUCAAUAGAUCCUUCAAUAAUAAGAUCUGAAGGCUUUCCAAAGGGUGUGCAUACAGAGCAUUCUUUUCUAUCCGAUAGUACAGUUAGUGUGUCG ..((((((((((....((-((..........((((((............))))))...((...((....))...)).....))))...))))))))))(((((.(((.....)))))))) ( -27.10, z-score = -1.32, R) >anoGam1.chr2L 39024711 119 + 48795086 GGGCCAGCAGCACGCAAU-CGACGCCGGCAAUCUUUCCCUCGAUCAGCACAUCGGACGGAAUGCCAAAGUGUGUGUUUACCACGCGCUCGGUGCGAUUCUCGAUUAUCUGACUAUCGUCC ((((.(((((..((.(((-((.((((((((.(((.(((...(((......)))))).))).))))..((((((((.....)))))))).)))))))))..)).....))).))...)))) ( -38.60, z-score = -0.95, R) >triCas2.chrUn_53 426484 119 + 446649 GGGCCAAGAGCACGCAUU-CAACCCCCUCGAUUUGGCCCAAAGUUAACACGUCGGAAGGCUUGCCGAAGGGCGUAUUGACCGACUUCGUUUUAGCGUUUUUUAAAAUGUCGGGGUUGUCG ((((((((((........-.......)))...)))))))...(((((.(((((....)))..(((....))))).)))))((((((((.....((((((....))))))))))))))... ( -38.46, z-score = -0.95, R) >consensus GCGAUAGCAG_GCACACU_UGACGCCUUCUAUCUGGCCAUCAAUGAUCACAUCGGAAGGUUUUCCAAACGGCGUGCAUACAGCAUACUCCAUGCGAUCCGCUAGUAAGAUGGGUGUGUCC .....................(((((........(((((((.............)))))))........))))).............................................. (-10.26 = -10.40 + 0.14)

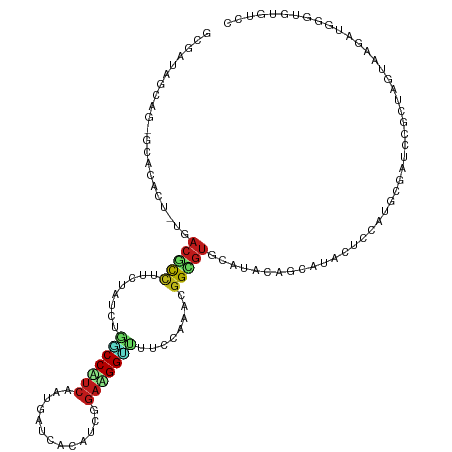
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:13 2011