
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,585,640 – 8,585,746 |
| Length | 106 |
| Max. P | 0.733460 |

| Location | 8,585,640 – 8,585,746 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Shannon entropy | 0.34765 |
| G+C content | 0.35953 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.733460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8585640 106 + 23011544 ----GUAUCUGUCUCUGUCUU-CCUCCUGGA-UUUUAUGUAAAUUUUU-CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGCU-GGAACCGGAGUGG ----.................-.((((.((.-(((((.(((.((....-.(..((((((((.(((((......))))).))))))))..)...)).))))-))))))))))... ( -23.80, z-score = -2.31, R) >droSim1.chr2L 8374934 106 + 22036055 ----GUAUCUGUCUCUGUCUU-CCUCCUGGA-UUUUAUGUAAAUUUUU-CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGCU-GGAACCGGAGUGG ----.................-.((((.((.-(((((.(((.((....-.(..((((((((.(((((......))))).))))))))..)...)).))))-))))))))))... ( -23.80, z-score = -2.31, R) >droSec1.super_3 4072542 106 + 7220098 ----GUAUCUGUCUCUGUCUU-CCUCCUGGA-UUUUAUGUAAAUUUUU-CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGCU-GGAACCGGAGUGG ----.................-.((((.((.-(((((.(((.((....-.(..((((((((.(((((......))))).))))))))..)...)).))))-))))))))))... ( -23.80, z-score = -2.31, R) >dp4.chr4_group5 1344064 108 - 2436548 ------AUCCCUUUGGCUUUUGCCAAACGGAGUUUUAUGCAAAUUUUUGCGUGCAAAUUGGUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGUUUGAAAUUGGAGUGC ------.(((.((((((....)))))).))).....((((((....))))))(((..(..(((((((.((((...((((((.....))))))...)))))))))))..)..))) ( -32.30, z-score = -3.28, R) >droPer1.super_5 1314700 108 - 6813705 ------AUCCCUUUGGCUUUUGGCAAACGGAGUUUUAUGCAAAUUUUUGCGUGCAAAUUGGUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGUUUGAAAUUGGAGUGC ------.(((.((((.(....).)))).))).....((((((....))))))(((..(..(((((((.((((...((((((.....))))))...)))))))))))..)..))) ( -27.70, z-score = -1.56, R) >droWil1.scaffold_181071 1005787 98 + 1211509 --------CCAACUUUGGUGU-GCUCCC----UUUUAUGCAAAUUUU--CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGUU-GGAAUUGAAGUGG --------(((.(((..((..-..(((.----...((((((((....--.(..((((((((.(((((......))))).))))))))..)))))))))..-)))))..)))))) ( -27.30, z-score = -3.33, R) >droVir3.scaffold_12963 14421903 106 - 20206255 ----CCCUCUGUCUCACUCCU-CAUCGUUUACUUUUAUGCAAAUUUU--CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUACGUU-GGAAUUGAAGUGG ----.((.((.((....(((.-...(((..........(((((....--.(..((((((((.(((((......))))).))))))))..)))))))))..-)))...)))).)) ( -20.60, z-score = -1.38, R) >droMoj3.scaffold_6500 7443070 110 - 32352404 CUUGGCAUCUGUCUCGCUCGU-CAGCGUUUACUUUUAUGCAAAUUUU--CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGCGUUUUGUACGUU-GGAAUUGAAGUGG ...(((....))).(((((((-((((((..((....(((((((((..--(((.((((.(((.......))).)))).))).)))))))))...)))))))-))...)).)))). ( -27.40, z-score = -1.19, R) >consensus ____G_AUCUGUCUCUCUCUU_CCUCCUGGA_UUUUAUGCAAAUUUUU_CGUGCAAAUUGCUUUUAAUGCAUUUUGAAUGCAAUUUGUGUUUUGUAUGUU_GGAAUUGGAGUGG ...................................((((((((.......(..((((((((.(((((......))))).))))))))..)))))))))................ (-13.46 = -13.72 + 0.27)
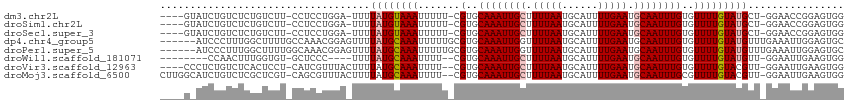

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:24 2011