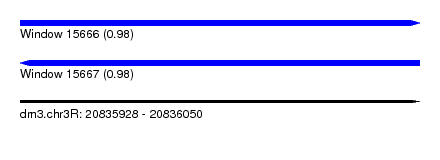
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,835,928 – 20,836,050 |
| Length | 122 |
| Max. P | 0.982963 |

| Location | 20,835,928 – 20,836,050 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 135 |
| Reading direction | forward |
| Mean pairwise identity | 71.64 |
| Shannon entropy | 0.51089 |
| G+C content | 0.34499 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.19 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
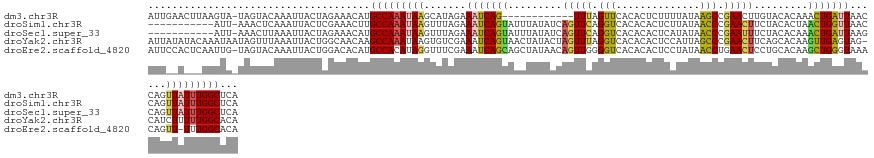
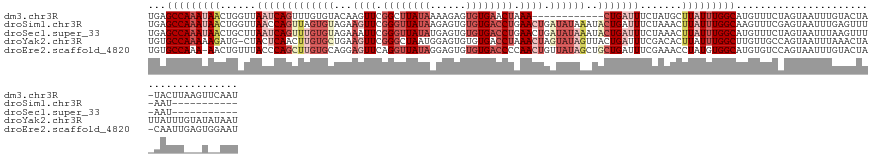
>dm3.chr3R 20835928 122 + 27905053 AUUGAACUUAAGUA-UAGUACAAAUUACUAGAAACAUGCCAAAUAAGCAUAGAAAUCAG------------UUUAGUUCACACUCUUUUAUAAGCCGAACUUGUACACAAACUGAUUAACCAGUUAUUUGGCUCA ...........((.-(((((.....)))))...))..(((((((((.......((((((------------((((((....)))....((((((.....))))))...)))))))))......)))))))))... ( -22.52, z-score = -1.72, R) >droSim1.chr3R 20667504 123 + 27517382 -----------AUU-AAACUCAAAUUACUCGAAACUUGCCAAAUAAGUUUAGAAAUCAGUAUUUAUAUCAGUUCAGGUCACACACUCUUAUAACCCGAACUUCUACACUAACUGGUUAACCAGUUAUUUGGCUCA -----------...-......................(((((((((((.((((................(((((.(((..............))).))))))))).))).(((((....)))))))))))))... ( -24.53, z-score = -3.32, R) >droSec1.super_33 97420 123 + 478481 -----------AUU-AAACUUAAAUUACUAGAAACAUGCCAAAUAAGUUUAGAAAUCAGUAUUUAUAUCAGUUCAGGUCACACACUCAUAUAACCCGAAUUUCUACACAAACUGAUUAAGCAGUUAUUUGGCUCA -----------...-......................((((((((((((((...((((((.........(((((.(((..............))).))))).........)))))))))))..)))))))))... ( -22.01, z-score = -2.76, R) >droYak2.chr3R 3169687 134 - 28832112 AUUAUAUACAAAUAAUAGUUUAAAUUACUGGCAACAAGCCAAAUAAGUGUCGAAAUCAGUAACUAUACUAGUUUAGGUCACACACUCCAUUAGCCCGAACUUCAGCACAAGUUGAGUAG-CAUCUUUUUGGCACA ............................((((.....)))).....(((((((((..((...((((...((((..(((..............)))..))))(((((....)))))))))-...))))))))))). ( -26.54, z-score = -1.79, R) >droEre2.scaffold_4820 3252696 133 - 10470090 AUUCCACUCAAUUG-UAGUACAAAUUACUGGACACAUGCCACAUAGGUUUCGAAAUCAGCAGCUAUAACAGUUGGGGUCACACACUCCUAUAACCUGAACUCCUGCACAAGCUGGGUAAACAGUU-UUUGGCACA ..((((.....(((-.....))).....))))....(((((....((((....)))).((((......(((..(((((.....)))))......))).....))))..((((((......)))))-).))))).. ( -25.90, z-score = 0.94, R) >consensus AUU__A___AAAUA_UAGUUCAAAUUACUAGAAACAUGCCAAAUAAGUUUAGAAAUCAGUAUCUAUAUCAGUUCAGGUCACACACUCCUAUAACCCGAACUUCUACACAAACUGAUUAACCAGUUAUUUGGCUCA .....................................(((((((((.......(((((((.........(((((.(((..............))).))))).........)))))))......)))))))))... (-15.75 = -16.19 + 0.44)
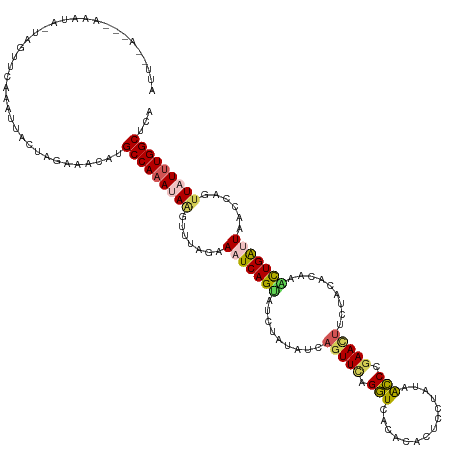
| Location | 20,835,928 – 20,836,050 |
|---|---|
| Length | 122 |
| Sequences | 5 |
| Columns | 135 |
| Reading direction | reverse |
| Mean pairwise identity | 71.64 |
| Shannon entropy | 0.51089 |
| G+C content | 0.34499 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -17.03 |
| Energy contribution | -20.55 |
| Covariance contribution | 3.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20835928 122 - 27905053 UGAGCCAAAUAACUGGUUAAUCAGUUUGUGUACAAGUUCGGCUUAUAAAAGAGUGUGAACUAAA------------CUGAUUUCUAUGCUUAUUUGGCAUGUUUCUAGUAAUUUGUACUA-UACUUAAGUUCAAU ((((((((((((((((..(((((((((.......((((((((((......)))).)))))))))------------)))))).))).).))))))))).......(((((.....)))))-.........))).. ( -29.81, z-score = -2.32, R) >droSim1.chr3R 20667504 123 - 27517382 UGAGCCAAAUAACUGGUUAACCAGUUAGUGUAGAAGUUCGGGUUAUAAGAGUGUGUGACCUGAACUGAUAUAAAUACUGAUUUCUAAACUUAUUUGGCAAGUUUCGAGUAAUUUGAGUUU-AAU----------- ...(((((((((((((....))))))).((((..(((((((((((((......)))))))))))))..))))....................))))))....((((((...))))))...-...----------- ( -37.40, z-score = -4.66, R) >droSec1.super_33 97420 123 - 478481 UGAGCCAAAUAACUGCUUAAUCAGUUUGUGUAGAAAUUCGGGUUAUAUGAGUGUGUGACCUGAACUGAUAUAAAUACUGAUUUCUAAACUUAUUUGGCAUGUUUCUAGUAAUUUAAGUUU-AAU----------- ...(((((((((......(((((((...((((..(.((((((((((((....)))))))))))).)..))))...))))))).......)))))))))......................-...----------- ( -36.02, z-score = -4.90, R) >droYak2.chr3R 3169687 134 + 28832112 UGUGCCAAAAAGAUG-CUACUCAACUUGUGCUGAAGUUCGGGCUAAUGGAGUGUGUGACCUAAACUAGUAUAGUUACUGAUUUCGACACUUAUUUGGCUUGUUGCCAGUAAUUUAAACUAUUAUUUGUAUAUAAU (((((....(((.((-.....)).))).(((((.....((((((((..((((((.(((....((.(((((....))))).)))))))))))..))))))))....)))))................))))).... ( -24.50, z-score = 0.86, R) >droEre2.scaffold_4820 3252696 133 + 10470090 UGUGCCAAA-AACUGUUUACCCAGCUUGUGCAGGAGUUCAGGUUAUAGGAGUGUGUGACCCCAACUGUUAUAGCUGCUGAUUUCGAAACCUAUGUGGCAUGUGUCCAGUAAUUUGUACUA-CAAUUGAGUGGAAU .((((((..-....((((...((((..((..((.((((..(((((((......)))))))..)))).))...)).))))......)))).....))))))...((((.(...(((.....-)))...).)))).. ( -32.50, z-score = 0.16, R) >consensus UGAGCCAAAUAACUGGUUAAUCAGUUUGUGUAGAAGUUCGGGUUAUAAGAGUGUGUGACCUAAACUGAUAUAAAUACUGAUUUCUAAACUUAUUUGGCAUGUUUCCAGUAAUUUGAACUA_AAUUU___U__AAU ...(((((((((......(((((((.........((((.((((((((......)))))))).)))).........))))))).......)))))))))..................................... (-17.03 = -20.55 + 3.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:11 2011