
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,830,417 – 20,830,513 |
| Length | 96 |
| Max. P | 0.878858 |

| Location | 20,830,417 – 20,830,513 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 61.95 |
| Shannon entropy | 0.69774 |
| G+C content | 0.43658 |
| Mean single sequence MFE | -14.08 |
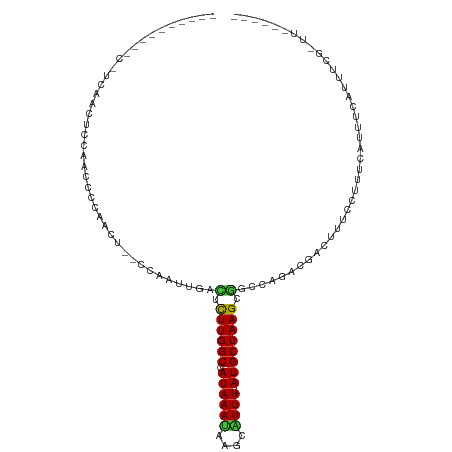
| Consensus MFE | -6.03 |
| Energy contribution | -5.40 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
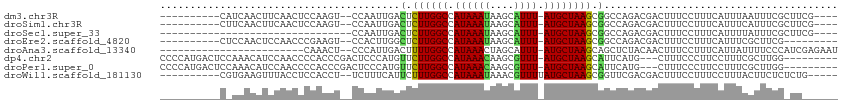
>dm3.chr3R 20830417 96 + 27905053 ----------CAUCAACUUCAACUCCAAGU--CCAAUUGACUCUUGGCCAUAAAUAAGCAUUU-AUGCUAAGCGGCCAGACGACUUUCCUUUCAUUUAAUUUCGCUUCG---- ----------................((((--(...(((.((((((((.((((((....))))-)))))))).)).)))..))))).......................---- ( -14.40, z-score = -1.08, R) >droSim1.chr3R 20661755 96 + 27517382 ----------CUUCAACUUCAACUCCAAGU--CCAAUUGACUCUUGGCCAUAAAUAAGCAUUU-AUGCUAAGCGGCCAGACGACUUUCCUUUCAUUUCAUUUCGCUUCG---- ----------................((((--(...(((.((((((((.((((((....))))-)))))))).)).)))..))))).......................---- ( -14.40, z-score = -1.07, R) >droSec1.super_33 91651 76 + 478481 --------------------------------CCAAUUGACUCUUGGCCAUAAAUAAGCAUUU-AUGCUAAGCGGCCAGACGACUUUCCUUUCAUUUUAUUUCGCUUCG---- --------------------------------...........((((((.......((((...-.))))....))))))..............................---- ( -10.60, z-score = -0.12, R) >droEre2.scaffold_4820 3247312 91 - 10470090 ----------CUCCAACUCCAACCCGAAGU--CCACUUGGCUCUUGGCCAUAAAUAAGCAUUU-AUGCUAAGCGGCCAGACGACUUUCCUUUCAUUUCGCUUCG--------- ----------...............(((((--(...((((((((((((.((((((....))))-)))))))).))))))..)))))).................--------- ( -22.80, z-score = -3.31, R) >droAna3.scaffold_13340 4872018 86 - 23697760 ------------------------CAAACU--CCCAUUGACUUUUGGCCAUAAACUAGCAUUU-AUGCUAAGCAGCUCUACAACUUUCCUUUCAUUAUUUUCCCAUCGAGAAU ------------------------....((--((((........)))........(((((...-.))))).....................................)))... ( -8.20, z-score = -0.06, R) >dp4.chr2 19889248 100 - 30794189 CCCCAUGACUCCAAACAUCCAACCCCACCCGACUCCCAUGUUCUUGGCCAUAAACAAGCGUUU-AUGCUAAGCAUUCAUG---CUUUCCCUUCCUUUCGCUUGG--------- ........................(((..(((....((((..((((((.((((((....))))-))))))))....))))---.............)))..)))--------- ( -15.23, z-score = -1.76, R) >droPer1.super_0 9562750 100 + 11822988 CCCCAUGACUCCAAACAUCCAACCCCACCCGACUCCCAUGUUCUUGGCCAUAAACAAGCGUUU-AUGCUAAGCAUUCAUG---CUUUCCCUUCCUUUCGCUUGG--------- ........................(((..(((....((((..((((((.((((((....))))-))))))))....))))---.............)))..)))--------- ( -15.23, z-score = -1.76, R) >droWil1.scaffold_181130 4067351 96 - 16660200 ----------CGUGAAGUUUACCUCCACCU--UCUUUCAUUCUUUGGCCAUAAAUAAACGUUUUAUGCUAAGCGGUUCGACGACUUUCCUUUCCUUUACUUCUCUCUG----- ----------.((((((.............--..........((((((.(((((.......)))))))))))((......))...........)))))).........----- ( -11.80, z-score = -0.55, R) >consensus __________C_UCAACUCCAACCCCAACU__CCAAUUGACUCUUGGCCAUAAAUAAGCAUUU_AUGCUAAGCGGCCAGACGACUUUCCUUUCAUUUCAUUUCG_UU______ ........................................(.((((((...((((....))))...)))))).)....................................... ( -6.03 = -5.40 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:10 2011