| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,830,108 – 20,830,206 |
| Length | 98 |
| Max. P | 0.781796 |

| Location | 20,830,108 – 20,830,206 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Shannon entropy | 0.43273 |
| G+C content | 0.53771 |
| Mean single sequence MFE | -31.49 |
| Consensus MFE | -15.75 |
| Energy contribution | -15.83 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20830108 98 - 27905053 -GGGUUCGUGCGGG-----UUGGCCCGCAAGUUCGUGAUCUCCGGCUC-GCGUACUUGUUCUCUAAGUGGCUACGAAAUUCCACAGCCACGCAGCCAACACAUCU----------- -......(((..((-----(((....((((((.(((((.(....).))-))).)))))).......((((((..(......)..)))))).)))))..)))....----------- ( -33.50, z-score = -1.54, R) >droSim1.chr3R 20661460 98 - 27517382 -GGGUUCGUGCGGG-----UUGGCCCGCAAGUUCGUGAUCUCCGGCUC-GCGUACUUGUUCUCUAAGUGGCUACGAAAUUCCACAGCCACGCAGCCAACACAUCU----------- -......(((..((-----(((....((((((.(((((.(....).))-))).)))))).......((((((..(......)..)))))).)))))..)))....----------- ( -33.50, z-score = -1.54, R) >droSec1.super_33 91406 98 - 478481 -GGGUUCGUGCGGG-----UUGGCCCGCAAGUUCGUGAUCUCCGGCUC-GCGUACUUGUUCUCUAAGUGGAUACGAAAUUCCACAGCCACGCAGCCAACACAUCU----------- -.((((((((((((-----....)))((((((.(((((.(....).))-))).)))))).......(((((........))))).).)))).)))).........----------- ( -34.90, z-score = -2.10, R) >droYak2.chr3R 3163851 98 + 28832112 -GGGUUCGUGCGGG-----UUGGCCCGCAAGUUCGUGAUCUCCGGCUC-GCGUACUUGUUCUCUAAGUGGCUACGAAAUUCCACAGCCACGCAGGCAACACAUCU----------- -..((((.((((((-----....)))((((((.(((((.(....).))-))).)))))).......((((((..(......)..))))))))).).)))......----------- ( -33.20, z-score = -1.23, R) >droEre2.scaffold_4820 3247041 98 + 10470090 -GGGUUCGUGCGGG-----UUGGCCCGCAAGUUCGUGAUCUCCGGCUC-GCGUACUUGUUCUCUAAGUGGCUACGAAAUUCCGCAGCCACGCAGCCAACACAUCU----------- -......(((..((-----(((....((((((.(((((.(....).))-))).)))))).......((((((.((......)).)))))).)))))..)))....----------- ( -37.20, z-score = -2.28, R) >dp4.chr2 19889012 111 + 30794189 UGGCUUGCCCCGCG-----GUAACCCGCAAGUUCGUGAUCUCUAACUCUGCGUACUUGUUCUCUAAGUGGCUAUGAAAUUCCUCAGCCACGCAGCCAACACAUCUUCGGCAUCGCC .(((.((((..(((-----(....))))((((.(((((........)).))).))))(((..((..((((((..(......)..))))))..))..)))........))))..))) ( -31.80, z-score = -2.01, R) >droPer1.super_0 9562533 92 - 11822988 UGGCUUGCCCCGCG-----GUAACCCGCAAGUUCGUGAUCUCUAACUCUGCGUACUUGUUCUCUAAGUGGCUAUGAAAUUCCUCAGCCACGCAGCCA------------------- .((((.((...(((-----(....))))((((.(((((........)).))).)))).........((((((..(......)..)))))))))))).------------------- ( -28.90, z-score = -2.69, R) >droWil1.scaffold_181130 4067104 103 + 16660200 UGGCUUCGUGCGGGCU-GCUUAACCCGCAAGUUCGUGAUCUCUAACUU-GCGUACUUGUUCUCUAAGUGGCUAUGAAAUUCCACAGUCACACACACACAGCGUCU----------- ..(((..(((((((..-......)))(((((((.(......).)))))-))))))...........((((((.((......)).))))))........)))....----------- ( -28.60, z-score = -1.33, R) >droVir3.scaffold_13047 12949677 96 - 19223366 ----------UGGGCU-GCUUAACCCGCAAGUUCGUGAUCUCUAACU-CGCGUAGUUGUUCUUCAAGUGGCUAUGAAAUUCCACAGCUACGCAGCCAACACACACACA-------- ----------..((((-((.......((((.(.(((((........)-)))).).)))).......((((((.((......)).))))))))))))............-------- ( -30.20, z-score = -3.02, R) >droMoj3.scaffold_6540 5243742 96 + 34148556 ----------UGGGCC-GCUUAACCCGCAAGUUCGUGAUCUCUAACU-CGCGUACUUGUUCUUCAAGUGGCUAUGAAAUUCCACAGCUACGCAGCGAGCACACAUAUA-------- ----------...(((-(((......((((((.(((((........)-)))).)))))).......((((((.((......)).))))))..)))).)).........-------- ( -29.00, z-score = -2.26, R) >droGri2.scaffold_15074 6013460 98 + 7742996 ----------UGGGCCAGCUUAACCCGCAAGUUCGUGAUCUCUAACUCCGCGUACUUGUUGCUCAAGUGGCUAUGAAAUUCCACAGCUACGCAGCCAACACACACACU-------- ----------..(((..((.......((((((.((((...........)))).)))))).......((((((.((......)).)))))))).)))............-------- ( -25.60, z-score = -1.29, R) >consensus _GGGUUCGUGCGGG_____UUAACCCGCAAGUUCGUGAUCUCUAACUC_GCGUACUUGUUCUCUAAGUGGCUAUGAAAUUCCACAGCCACGCAGCCAACACAUCU___________ ...........(((.........)))((((((.(((.............))).)))))).......((((((..(......)..)))))).......................... (-15.75 = -15.83 + 0.08)

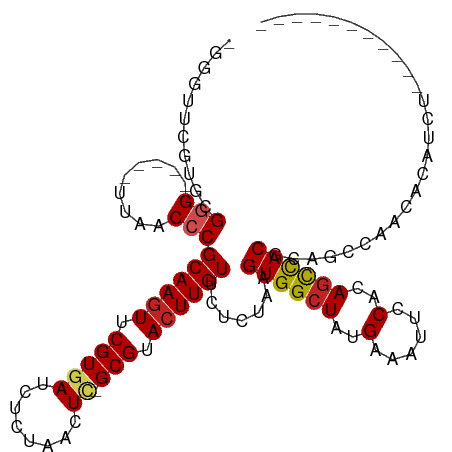

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:09 2011