
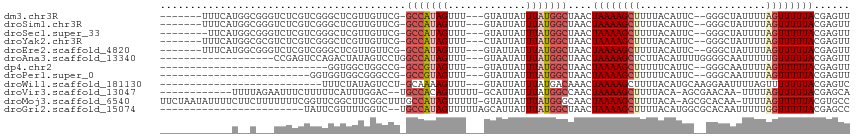
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,813,147 – 20,813,249 |
| Length | 102 |
| Max. P | 0.551539 |

| Location | 20,813,147 – 20,813,249 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.23 |
| Shannon entropy | 0.64337 |
| G+C content | 0.37155 |
| Mean single sequence MFE | -21.59 |
| Consensus MFE | -7.35 |
| Energy contribution | -7.81 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
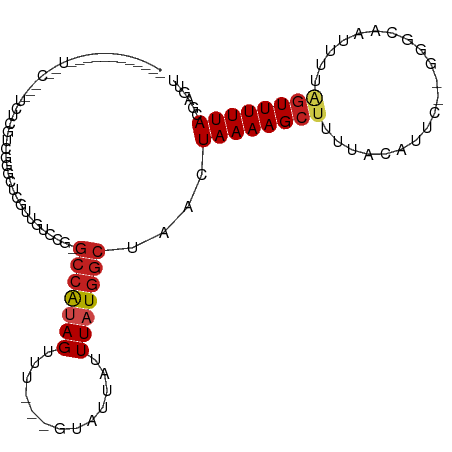
>dm3.chr3R 20813147 102 - 27905053 -------UUUCAUGGCGGGUCUCGUCGGGCUCGUUGUUCG-GCCAUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC--GGGCUAUUUUAGUUUUUACGAGUU -------....(..((((((((....))))))))..)(((-(((((((...---.......))))))).((((((((((((........--)))))..)))))))....)))... ( -28.50, z-score = -3.10, R) >droSim1.chr3R 20644795 102 - 27517382 -------UUUCAUGGCGGGUCUCGUCGGGCUCGUUGUUCG-GCCAUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC--GGGCUAUUUUAGUUUUUACGAGUU -------....(..((((((((....))))))))..)(((-(((((((...---.......))))))).((((((((((((........--)))))..)))))))....)))... ( -28.50, z-score = -3.10, R) >droSec1.super_33 74556 101 - 478481 --------UUCAUGGCGGGUCUCGUCGGGCUCGUUGUUCG-GCCAUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC--GGGCUAUUUUAGUUUUUACGAGUU --------...(..((((((((....))))))))..)(((-(((((((...---.......))))))).((((((((((((........--)))))..)))))))....)))... ( -28.50, z-score = -3.05, R) >droYak2.chr3R 3146333 102 + 28832112 -------UUUCAUGGCGCGUCUCGUCGGGCUCGUUGUUCG-GCCAUAGUUU---CUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC--GGGCUAUUUUAGUUUUUACGAGUU -------.....(((((.....)))))(((((((.....(-(((((((...---.......))))))))((((((((((((........--)))))..)))))))...))))))) ( -26.40, z-score = -2.46, R) >droEre2.scaffold_4820 3230196 102 + 10470090 -------UUUCAUGGCGGGUCUCGUCGGGCUCGUUGUUCG-GCCAUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC--GGGCUAUUUUAGUUUUUACGAGUU -------....(..((((((((....))))))))..)(((-(((((((...---.......))))))).((((((((((((........--)))))..)))))))....)))... ( -28.50, z-score = -3.10, R) >droAna3.scaffold_13340 4855925 93 + 23697760 -------------------CCGAGUCCAGACUAUAGUCCUGGCCAUAGUUU---GUAAUAUUUAUGGCUAACUAAAAGCUCUUACAUUUUGGGGCAAUUUUUGUUUUUACGAGUU -------------------....((..((((..((((..(((((((((...---.......)))))))))))))...(((((........))))).......))))..))..... ( -20.40, z-score = -0.93, R) >dp4.chr2 19869488 81 + 30794189 ----------------------------GGUGGCUGGCCG-GCCGUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUUCAUUC--GGGCAAUUUUAGUUUUUACGAGUU ----------------------------(((.....)))(-(((((((...---.......))))))))((((((((((((........--))))..)))))))).......... ( -17.00, z-score = -0.00, R) >droPer1.super_0 9542859 84 - 11822988 -------------------------GGUGGUGGCGGGCCG-GCCGUAGUUU---GUAUUAUUUAUGGCUAACUAAAAGCUUUUUCAUUC--GGGCAAUUUUAGUUUUUACGAGUU -------------------------.((((.((....))(-(((((((...---.......))))))))((((((((((((........--))))..)))))))).))))..... ( -17.30, z-score = 0.37, R) >droWil1.scaffold_181130 4049280 84 + 16660200 ---------------------------UUUCUAUAGUCCU-GCAAAAGUUU---GUAUUAUUUAUGACAAACUAAAAGCUUUUACAUGCAAGGAAUUUUAGUUUUUUUACGAGUC ---------------------------...(((...((((-.....(((((---((..........)))))))....((........)).))))....))).............. ( -12.20, z-score = -0.25, R) >droVir3.scaffold_13047 12931259 98 - 19223366 ------------UUUUAGAAUUUCUUUUUCAUUUGGAC--UGCCACAGUUUUU-GCAUUAUUUAUGGCCAACUAAAAGCUUUUACA-AGCGAACAA-UUUUAGUUUUUACGAGCA ------------.....(((.......))).(((((..--.((((.((.....-.......)).))))...))))).((((.((.(-(((......-.....)))).)).)))). ( -12.20, z-score = 0.40, R) >droMoj3.scaffold_6540 5225491 112 + 34148556 UUCUAAUAUUUUCUUCUUUUUUUCGGUUCGGCUUCGGCUUUGCCAUAGUUUUU-GUAUUAUUUAUGGGCAACUAAAAGCUUUUACA-AGCGCACAA-UUUUAGUUUUUACGUGCC ........................(((..(((....)))..))).(((((((.-.((.....))..)).)))))...((((....)-)))((((..-.............)))). ( -21.16, z-score = -1.48, R) >droGri2.scaffold_15074 5991192 89 + 7742996 ------------------------UAUUCGUUUUGGUC--UGCCAUAGUUUUUAGCAUUAUUUAUGGCUAACUAAAAGCUUUUACAUGGCGCACAAUUUUUGGUUUUUACGAGCC ------------------------..(((((((((((.--.(((((((.............)))))))..)))))).(((.......)))..................))))).. ( -18.42, z-score = -1.17, R) >consensus ____________U__C___UCUCGUCGGGCUCGUUGUCCG_GCCAUAGUUU___GUAUUAUUUAUGGCUAACUAAAAGCUUUUACAUUC__GGGCAAUUUUAGUUUUUACGAGUU .........................................(((((((.............)))))))....((((((((.....................))))))))...... ( -7.35 = -7.81 + 0.47)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:05 2011