
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,812,700 – 20,812,779 |
| Length | 79 |
| Max. P | 0.990806 |

| Location | 20,812,700 – 20,812,779 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Shannon entropy | 0.31573 |
| G+C content | 0.48169 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -21.75 |
| Energy contribution | -22.39 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.965773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
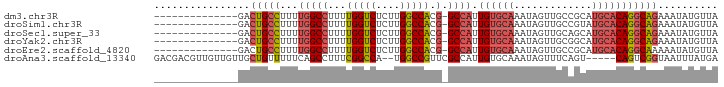
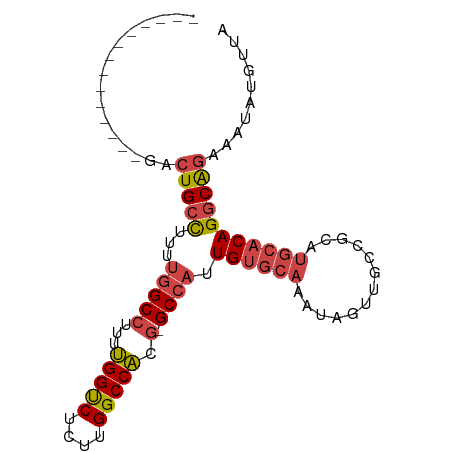
>dm3.chr3R 20812700 79 + 27905053 --------------GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG-GCCAUUGUGCAAAUAGUUGCCGCAUGCACAGGCAGAAAUAUGUUA --------------..(((((...(((((...(((((....))))).)-)))).((((((.............))))))))))).......... ( -30.02, z-score = -2.53, R) >droSim1.chr3R 20644384 79 + 27517382 --------------GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG-GCCAUUGUGCAAAUAGUUGCCGUAUGCACAGGCAGAAAUAUGUUA --------------..(((((...(((((...(((((....))))).)-)))).((((((.((.......)).))))))))))).......... ( -30.60, z-score = -3.08, R) >droSec1.super_33 74108 79 + 478481 --------------GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG-GCCAUUGUGCAAAUAGUUGCAGCAUGCACAGGCAGAAAUAUGUUA --------------..(((((...(((((...(((((....))))).)-)))).((((((.............))))))))))).......... ( -30.02, z-score = -2.42, R) >droYak2.chr3R 3145889 79 - 28832112 --------------GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG-GCCAUUGUGCAAAUAGUUGCGGCAUGCACAGGCAGAAAUAUGUUA --------------..(((((...(((((...(((((....))))).)-)))).((((((.............))))))))))).......... ( -30.02, z-score = -2.30, R) >droEre2.scaffold_4820 3229747 79 - 10470090 --------------GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG-GCCAUUGUGCAAAUAGUUGCCGCAUGCACAGGCAAAAAUAUGUUA --------------...((((...(((((...(((((....))))).)-)))).((((((.............))))))))))........... ( -26.72, z-score = -1.65, R) >droAna3.scaffold_13340 4855682 87 - 23697760 GACGACGUUGUUGUUGCUGUUUUUCAGCCUUUCGGCCA--UGGCCGUUCGCCAUUGUGCAAAUAGUUUCAGU-----CAGUCGGUAAUUUAUGA ..((((....(((..(((((((..(((((....))).(--((((.....)))))))...)))))))..))).-----..))))........... ( -22.00, z-score = -0.94, R) >consensus ______________GACUGCCUUUUGGCCUUUUGGUCUCUUGGCCACG_GCCAUUGUGCAAAUAGUUGCCGCAUGCACAGGCAGAAAUAUGUUA ................(((((...((((....(((((....)))))...)))).((((((.............))))))))))).......... (-21.75 = -22.39 + 0.64)
| Location | 20,812,700 – 20,812,779 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Shannon entropy | 0.31573 |
| G+C content | 0.48169 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.990806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
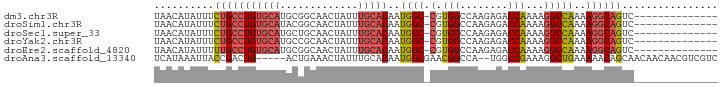
>dm3.chr3R 20812700 79 - 27905053 UAACAUAUUUCUGCCUGUGCAUGCGGCAACUAUUUGCACAAUGGC-CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC-------------- ..........(((((((((((...(....)....)))))..((((-(.(((........)))...)))))..))))))..-------------- ( -30.10, z-score = -3.20, R) >droSim1.chr3R 20644384 79 - 27517382 UAACAUAUUUCUGCCUGUGCAUACGGCAACUAUUUGCACAAUGGC-CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC-------------- ..........(((((((((((...(....)....)))))..((((-(.(((........)))...)))))..))))))..-------------- ( -30.10, z-score = -3.84, R) >droSec1.super_33 74108 79 - 478481 UAACAUAUUUCUGCCUGUGCAUGCUGCAACUAUUUGCACAAUGGC-CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC-------------- ..........(((((((((((.............)))))..((((-(.(((........)))...)))))..))))))..-------------- ( -27.82, z-score = -2.58, R) >droYak2.chr3R 3145889 79 + 28832112 UAACAUAUUUCUGCCUGUGCAUGCCGCAACUAUUUGCACAAUGGC-CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC-------------- ..........(((((((((((.............)))))..((((-(.(((........)))...)))))..))))))..-------------- ( -27.82, z-score = -2.76, R) >droEre2.scaffold_4820 3229747 79 + 10470090 UAACAUAUUUUUGCCUGUGCAUGCGGCAACUAUUUGCACAAUGGC-CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC-------------- ..........(((((((((((...(....)....)))))..((((-(.(((........)))...)))))..))))))..-------------- ( -28.20, z-score = -2.57, R) >droAna3.scaffold_13340 4855682 87 + 23697760 UCAUAAAUUACCGACUG-----ACUGAAACUAUUUGCACAAUGGCGAACGGCCA--UGGCCGAAAGGCUGAAAAACAGCAACAACAACGUCGUC ............(((.(-----((.........((((...(((((.....))))--)((((....))))........)))).......)))))) ( -22.29, z-score = -2.66, R) >consensus UAACAUAUUUCUGCCUGUGCAUGCGGCAACUAUUUGCACAAUGGC_CGUGGCCAAGAGACCAAAAGGCCAAAAGGCAGUC______________ ..........(((((((((((.............)))))..((((...(((........)))....))))..))))))................ (-18.28 = -18.75 + 0.47)
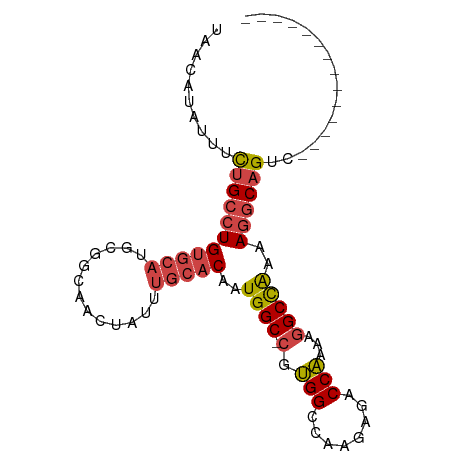
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:03 2011