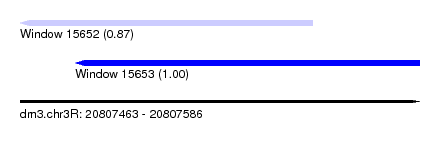
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,807,463 – 20,807,586 |
| Length | 123 |
| Max. P | 0.997984 |

| Location | 20,807,463 – 20,807,553 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Shannon entropy | 0.15681 |
| G+C content | 0.57662 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -28.98 |
| Energy contribution | -29.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
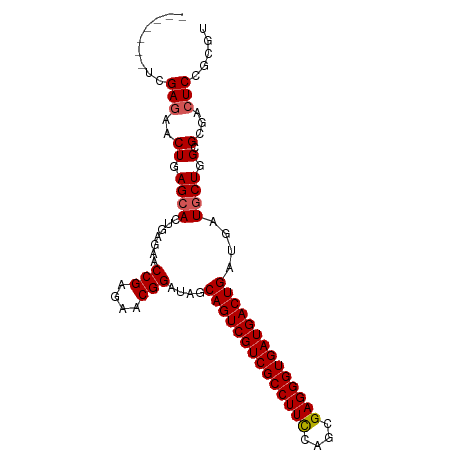
>dm3.chr3R 20807463 90 - 27905053 -------UCGAGAACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCCGCGU -------..........((((.......(((....)))....((((((((((((((....))))))))))))))....)))).(((((....))))) ( -36.10, z-score = -2.55, R) >droSim1.chr3R 20639090 90 - 27517382 -------UCGAGAACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGUCUCCGCGU -------..........((((.......(((....)))....((((((((((((((....))))))))))))))....)))).(((((....))))) ( -35.60, z-score = -1.92, R) >droSec1.super_33 68818 90 - 478481 -------UCGAGAACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGUCUCCGCGU -------..........((((.......(((....)))....((((((((((((((....))))))))))))))....)))).(((((....))))) ( -35.60, z-score = -1.92, R) >droYak2.chr3R 3140427 94 + 28832112 UCGAGAACUGAACACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUUCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCGU--- .((((..(........)((((.......(((....)))....((((((((((((((....))))))))))))))....)))).......)))).--- ( -30.30, z-score = -1.00, R) >droEre2.scaffold_4820 3224406 87 + 10470090 -------UCGAGAACUGAGAACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCGU--- -------.((((..((.(....).))...((....(((((..((((((((((((((....))))))))))))))...)).)))...)).)))).--- ( -31.60, z-score = -2.23, R) >consensus _______UCGAGAACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUGCUGGCGCGACUCCGCGU .........(((..((.((((.......(((....)))....((((((((((((((....))))))))))))))....)))).).)...)))..... (-28.98 = -29.22 + 0.24)

| Location | 20,807,480 – 20,807,586 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.13 |
| Shannon entropy | 0.08461 |
| G+C content | 0.52263 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -34.88 |
| Energy contribution | -34.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.997984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
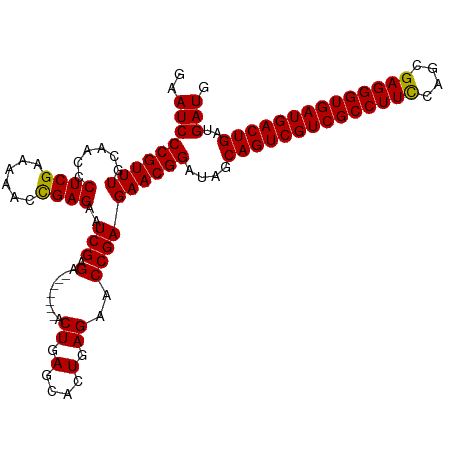
>dm3.chr3R 20807480 106 - 27905053 GAAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGA-------ACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUG ..(((((((((......((((......))))..(((.(.-------.((.(....).))..))))))))))....((((((((((((((....))))))))))))))..))). ( -36.40, z-score = -3.81, R) >droSim1.chr3R 20639107 106 - 27517382 GAAUCCCGUUUGCAACCCUCGAAAUAGCGAGAAUCGAGA-------ACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUG ..(((((((((......((((......))))..(((.(.-------.((.(....).))..))))))))))....((((((((((((((....))))))))))))))..))). ( -36.20, z-score = -3.33, R) >droSec1.super_33 68835 106 - 478481 GAAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGA-------ACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUG ..(((((((((......((((......))))..(((.(.-------.((.(....).))..))))))))))....((((((((((((((....))))))))))))))..))). ( -36.40, z-score = -3.81, R) >droYak2.chr3R 3140441 113 + 28832112 GAAUCCCGUUUGCAACCCUCGAAAAACUGAGAAUCGAGAACUGAACACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUUCAGCGAGGGUGAUGACUGAUGAUG ..((((((((..(....(((((...........))))).........((.(....).))....)..)))))....((((((((((((((....))))))))))))))..))). ( -33.20, z-score = -2.33, R) >droEre2.scaffold_4820 3224420 105 + 10470090 GAAUCCCGUUUGCAACCCUCGAAA-AGCGAGAAUCGAGA-------ACUGAGAACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUG ..(((((((((......((((...-..))))..(((.(.-------.((.(....).))..))))))))))....((((((((((((((....))))))))))))))..))). ( -36.00, z-score = -3.66, R) >consensus GAAUCCCGUUUGCAACCCUCGAAAAACCGAGAAUCGAGA_______ACUGAGCACUGAGAACCGAGAACGGAUAGCAGUCGUCGCCUUCCAGCGAGGGUGAUGACUGAUGAUG ..((((((((..(....(((((...........))))).........((.(....).))....)..)))))....((((((((((((((....))))))))))))))..))). (-34.88 = -34.72 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:42:00 2011