
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,779,234 – 20,779,363 |
| Length | 129 |
| Max. P | 0.657880 |

| Location | 20,779,234 – 20,779,328 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Shannon entropy | 0.26474 |
| G+C content | 0.61931 |
| Mean single sequence MFE | -24.54 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.642487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20779234 94 + 27905053 GCAGCUGCCCAUCCCCAUCCGCAUCCCCAUCCUCGUCUUUAGUCUGGCCCUCAACCUUGGCCACAGCCCCAGCAUUUGGCCAAAGACAGCAGCC ...(((((............(......)......((((((.((((((((.........)))))..((....))....))).)))))).))))). ( -23.00, z-score = -1.74, R) >droSim1.chr3R 20611137 93 + 27517382 GCAGCUGCCCAUCCCCACCC-CAUCACCAUCCUCGUCUUCGGUCUGGCUCUCAGCCUUGGCCCCAGCCCCAGCAUUUGGCCAAAGACAGCAGCC ...(((((............-............((....))(((((((.....)))((((((...((....))....)))))))))).))))). ( -25.90, z-score = -1.77, R) >droSec1.super_33 40783 88 + 478481 GCAGCUGCCCAUCCCCA------UCACCAUCCUCGUCUUCGGUCUGGCUCUCAGCCUUGGCCCCAGCCCCAGCAUUUGGCCAAAGACAGCAGCC ...(((((.........------..........((....))(((((((.....)))((((((...((....))....)))))))))).))))). ( -25.90, z-score = -1.63, R) >droYak2.chr3R 3111601 94 - 28832112 GCAGCUGCCCAUCCUCAUACUCCAGCUCAGCCUCGGUCUCAGUCUGGCUCUUAGCCUUGGCCACAUCCCCAGCAUUUGGCCAAAGACAGCAGCC ...(((((.............((((......)).)).....(((((((.....)))(((((((.((.......)).))))))))))).))))). ( -25.70, z-score = -0.67, R) >droEre2.scaffold_4820 3195357 88 - 10470090 GCAGCUGCCCAUCCUCA------GCCUCAUCCUUCACCUCAGUCCGGCUCUUAGCCUUGGCCACAUCCCCAGCAUUUGGCCAAAGACAGCAGCC ((.((((........))------))................(((.(((.....)))(((((((.((.......)).))))))).))).)).... ( -22.20, z-score = -1.47, R) >consensus GCAGCUGCCCAUCCCCA__C_C_UCCCCAUCCUCGUCUUCAGUCUGGCUCUCAGCCUUGGCCACAGCCCCAGCAUUUGGCCAAAGACAGCAGCC ...(((((..........................(....).(((((((.....)))((((((...............)))))))))).))))). (-19.28 = -19.76 + 0.48)

| Location | 20,779,234 – 20,779,328 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Shannon entropy | 0.26474 |
| G+C content | 0.61931 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.55 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.657880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
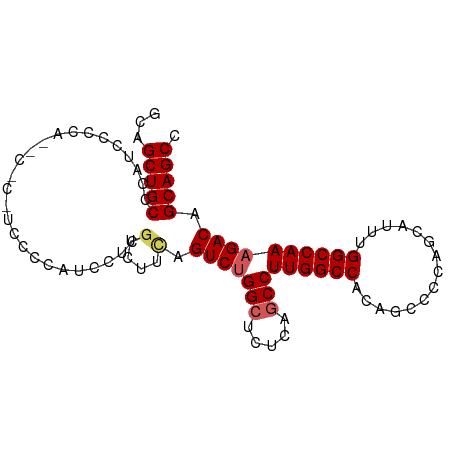
>dm3.chr3R 20779234 94 - 27905053 GGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGUGGCCAAGGUUGAGGGCCAGACUAAAGACGAGGAUGGGGAUGCGGAUGGGGAUGGGCAGCUGC (((((((((((((((((....(((..(((....)))..)))....))))).....)))))..(.((....)).)...........))))))).. ( -33.20, z-score = -1.08, R) >droSim1.chr3R 20611137 93 - 27517382 GGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGGGGCCAAGGCUGAGAGCCAGACCGAAGACGAGGAUGGUGAUG-GGGUGGGGAUGGGCAGCUGC (((((((((((..(.((....(((..(((....)))..))).....((((..(((....)).)..))))....-)).)..)))).))))))).. ( -36.70, z-score = -1.55, R) >droSec1.super_33 40783 88 - 478481 GGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGGGGCCAAGGCUGAGAGCCAGACCGAAGACGAGGAUGGUGA------UGGGGAUGGGCAGCUGC (((((((..((((((((....((....))...))))))))((.(..((((..(((....)).)..))))..------).))....))))))).. ( -36.20, z-score = -1.91, R) >droYak2.chr3R 3111601 94 + 28832112 GGCUGCUGUCUUUGGCCAAAUGCUGGGGAUGUGGCCAAGGCUAAGAGCCAGACUGAGACCGAGGCUGAGCUGGAGUAUGAGGAUGGGCAGCUGC (((((((..(((((((((....(....)...)))))))))((.....((((.((.((.(....))).))))))......))....))))))).. ( -35.30, z-score = -1.00, R) >droEre2.scaffold_4820 3195357 88 + 10470090 GGCUGCUGUCUUUGGCCAAAUGCUGGGGAUGUGGCCAAGGCUAAGAGCCGGACUGAGGUGAAGGAUGAGGC------UGAGGAUGGGCAGCUGC ((((((((((((..(((.....((((.....((((....))))....)))).((.......)).....)))------..))))).))))))).. ( -34.10, z-score = -2.05, R) >consensus GGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGUGGCCAAGGCUGAGAGCCAGACUGAAGACGAGGAUGGGGA_G_G__UGGGGAUGGGCAGCUGC ((((((...(((((((((...((....))..))))))))).......(((..((.........................))..))))))))).. (-27.39 = -27.55 + 0.16)



| Location | 20,779,269 – 20,779,363 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Shannon entropy | 0.18098 |
| G+C content | 0.57607 |
| Mean single sequence MFE | -37.68 |
| Consensus MFE | -30.94 |
| Energy contribution | -32.90 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
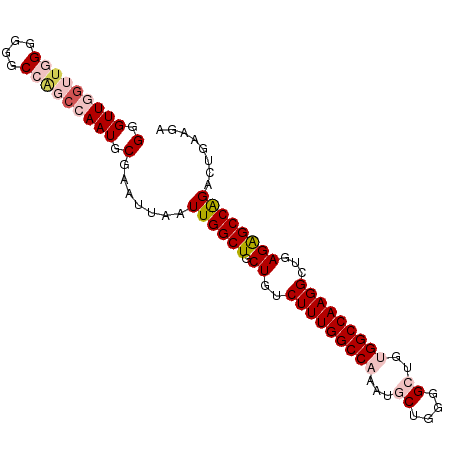
>dm3.chr3R 20779269 94 - 27905053 GGGUUGGUUGGGGGGCCAGCCAAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGUGGCCAAGGUUGAGGGCCAGACUAAAGA (.(((((((((....))))))))).).......((((((.((..(((((((((...((....))..)))))))))...))))))))........ ( -40.50, z-score = -2.27, R) >droSim1.chr3R 20611171 94 - 27517382 GGGUUGGUUGGGGGGCCAGCCAAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGGGGCCAAGGCUGAGAGCCAGACCGAAGA (((((((((((....))))))))).........((((((.((..((((((((....((....))...))))))))...)))))))).))..... ( -39.00, z-score = -1.05, R) >droSec1.super_33 40812 94 - 478481 GGGUUGGUUGGGGGGCCAGCCAAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGGGGCCAAGGCUGAGAGCCAGACCGAAGA (((((((((((....))))))))).........((((((.((..((((((((....((....))...))))))))...)))))))).))..... ( -39.00, z-score = -1.05, R) >droYak2.chr3R 3111636 89 + 28832112 GGGUUGGUUGGGGGCC-----AAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGAUGUGGCCAAGGCUAAGAGCCAGACUGAGAC (.((((((.....)))-----))).).......(((((..(.(((((..((.....))..))))))..)))))(((.....))).......... ( -33.40, z-score = -2.05, R) >droEre2.scaffold_4820 3195386 94 + 10470090 GGGUUGGUUGGGGGCCAGCCGAAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGAUGUGGCCAAGGCUAAGAGCCGGACUGAGGU .(((((((.....))))))).............(((((..(.(((((..((.....))..))))))..)))))(((.....))).......... ( -36.50, z-score = -1.51, R) >consensus GGGUUGGUUGGGGGGCCAGCCAAUGCGAAUUAAUUGGCUGCUGUCUUUGGCCAAAUGCUGGGGCUGUGGCCAAGGCUGAGAGCCAGACUGAAGA (.(((((((((....))))))))).).......((((((.((..(((((((((...((....))..)))))))))...))))))))........ (-30.94 = -32.90 + 1.96)

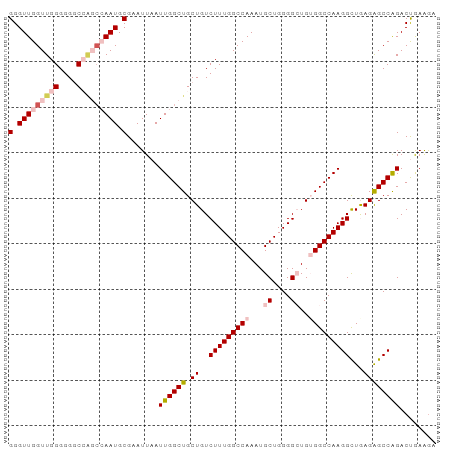
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:53 2011