
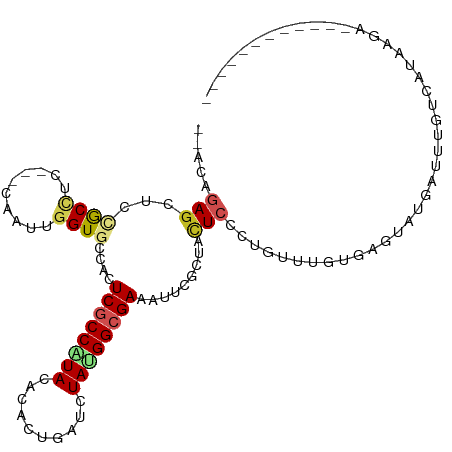
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,747,186 – 20,747,285 |
| Length | 99 |
| Max. P | 0.822769 |

| Location | 20,747,186 – 20,747,285 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.62303 |
| G+C content | 0.47517 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -9.28 |
| Energy contribution | -9.74 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20747186 99 + 27905053 --ACCGAGCGCAGCCUCCUCCAAUUGGUGCCACUCGCCAUACACAUUUAUCUAUGGCGAAAUUCGAUACUCUCUGUUUGUGAGUAUGAUUUGCCAUAAGUA----------- --..((((.(((.((..........)))))..))))...........((..(((((((((..((.((((((.(.....).)))))))))))))))))..))----------- ( -26.40, z-score = -2.29, R) >droGri2.scaffold_14624 1782054 109 - 4233967 GAGGAGAGUCCAGCUUC---GCAUUGGUGCCAUUCGCCAUAUACGCUUGUCUACGGCGAAAUACGCUACUCCGUUUUUGUGAGUACCAGCAGUGAUAAGCCCUAUACAAAGA ..((.....)).((((.---.(((((.((...((((((.((.((....)).)).))))))......(((((((....)).))))).)).)))))..))))............ ( -26.80, z-score = -0.07, R) >droVir3.scaffold_12855 2486972 95 + 10161210 --ACCGAGUCCGGCUUC---ACAUUGGUGCCAUUCGCCCUAUACGCUCGUCUACGGCGAAAUACGCUACUCCGUUUAUGUGUGU---AACUUUCUUAUG---UAUA------ --((.((((..(((.((---.....)).))).((((((.((.((....)).)).)))))).......)))).))...((((..(---((.....)))..---))))------ ( -20.80, z-score = -0.35, R) >droWil1.scaffold_181130 3990234 101 - 16660200 --GCAAAACUCUGCUUC---CAAUUGGUGUCACUCGCCCUAUACUCUGAUGUAUGGGGAAAUUCGUUACUCGGUCUUUGUAAGUCCAAAAGGCUGCAGCGCCUGCA------ --(((...(.((((..(---(....((((.......((((((((......))))))))........)))).((.((.....)).))....))..)))).)..))).------ ( -28.56, z-score = -1.66, R) >droPer1.super_0 9481356 93 + 11822988 --CCAGAGUUCCGCCUC---CCACUGGUGUCACUCGCCGUACACCCUUAUCUACGGCGAAAUACGGUACUCCCUGUUUGUAUGU-----UUGUCUGAAAAUCU--------- --.((((((..((((..---.....))))..))((((((((..........)))))))).((((((.((.....))))))))..-----...)))).......--------- ( -24.10, z-score = -2.32, R) >dp4.chr2 19808093 93 - 30794189 --CCAGAGUUCCGCCUC---CCACUGGUGUCACUCGCCGUACACCCUUAUCUACGGCGAAAUACGGUACUCCCUGUUUGUAUGU-----UUGUCUGAAAAUCA--------- --.((((((..((((..---.....))))..))((((((((..........)))))))).((((((.((.....))))))))..-----...)))).......--------- ( -24.10, z-score = -2.13, R) >droAna3.scaffold_13340 4795367 95 - 23697760 --GGAUAGCUCUACAUC---CAACUGGUGUCACUCACCGUAUACCUUAAUCUAUGGCGAGAUUAGGUAUUCUCUUUAUGUAAGUAUAAUUAGGAGAGGAA------------ --((((........)))---)....((((.....))))...(((((.(((((......))))))))))((((((((...(((......))))))))))).------------ ( -22.40, z-score = -0.49, R) >droEre2.scaffold_4820 3167723 93 - 10470090 --ACCGAGCUCCGCAUC---CAAUUGGUGCCACUCGCCAUACACACUGAUUUAUGGCGAAAUCCGCUAUUCCCUGUUUGUGAGUAUGAUUUGCCUUAA-------------- --..((((....(((((---.....)))))..)))).(((((.(((.(((..((((((.....)))))).....))).))).)))))...........-------------- ( -24.00, z-score = -2.39, R) >droYak2.chr3R 3084228 95 - 28832112 --ACAGAGCUCCGCCUC---CAACUGGUGCCACUCGCCAUACACACUGAUCUAUGGCGAAAUUCGUUAUUCUCUGUUUGUGAGUAUGAUUUGCCACACCA------------ --((((((...((((..---.....))))....((((((((..........))))))))...........)))))).((((.(((.....)))))))...------------ ( -25.60, z-score = -2.26, R) >droSec1.super_0 21094725 99 + 21120651 --ACCGAGCGCCGCCUCCUCCAAUUGGUGCCACUCGCCAUACACACUGAUCUAUGGCGAAAUUCGCUACUCUCUGUUUGUGAGUAUGAUUUGUCAUAAGUA----------- --..((((.(.((((..........)))).).))))...............(((((((((..(((.(((((.(.....).)))))))))))))))))....----------- ( -25.10, z-score = -1.60, R) >droSim1.chr3R 20582828 99 + 27517382 --ACCGAGCGCCGCCUCCUCCAAUUGGUGCCACUCGCCAUACACACUGAUCUAUGGCGAAAUUCGCUACUCUCUGUUUGUGAGUAUGAUUUGUCAUAAGUA----------- --..((((.(.((((..........)))).).))))...............(((((((((..(((.(((((.(.....).)))))))))))))))))....----------- ( -25.10, z-score = -1.60, R) >consensus __ACAGAGCUCCGCCUC___CAAUUGGUGCCACUCGCCAUACACACUGAUCUAUGGCGAAAUUCGCUACUCCCUGUUUGUGAGUAUGAUUUGUCAUAAGA____________ .....(((...((((..........))))....((((((((..........)))))))).........)))......................................... ( -9.28 = -9.74 + 0.46)


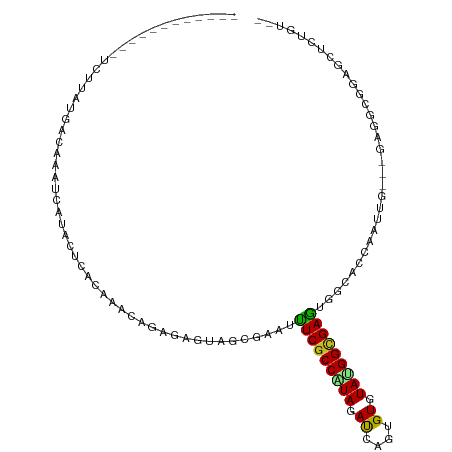
| Location | 20,747,186 – 20,747,285 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.62303 |
| G+C content | 0.47517 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -12.02 |
| Energy contribution | -11.71 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20747186 99 - 27905053 -----------UACUUAUGGCAAAUCAUACUCACAAACAGAGAGUAUCGAAUUUCGCCAUAGAUAAAUGUGUAUGGCGAGUGGCACCAAUUGGAGGAGGCUGCGCUCGGU-- -----------.(((...(((...((((((((.(.....).)))))).))...((((((((.((....)).))))))))(..((.((.......))..))..)))).)))-- ( -29.00, z-score = -1.90, R) >droGri2.scaffold_14624 1782054 109 + 4233967 UCUUUGUAUAGGGCUUAUCACUGCUGGUACUCACAAAAACGGAGUAGCGUAUUUCGCCGUAGACAAGCGUAUAUGGCGAAUGGCACCAAUGC---GAAGCUGGACUCUCCUC .((((((((.(((((......(((.(.(((((.(......)))))).)))).(((((((((.((....)).))))))))).))).)).))))---))))..((.....)).. ( -34.30, z-score = -1.86, R) >droVir3.scaffold_12855 2486972 95 - 10161210 ------UAUA---CAUAAGAAAGUU---ACACACAUAAACGGAGUAGCGUAUUUCGCCGUAGACGAGCGUAUAGGGCGAAUGGCACCAAUGU---GAAGCCGGACUCGGU-- ------..((---(((......(((---((.(.(......)).)))))((..((((((.((.((....)).)).))))))..))....))))---)..((((....))))-- ( -23.90, z-score = -0.90, R) >droWil1.scaffold_181130 3990234 101 + 16660200 ------UGCAGGCGCUGCAGCCUUUUGGACUUACAAAGACCGAGUAACGAAUUUCCCCAUACAUCAGAGUAUAGGGCGAGUGACACCAAUUG---GAAGCAGAGUUUUGC-- ------.((((((.((((..((..((((..((((......((.....))....(((((((((......)))).))).))))))..))))..)---)..)))).).)))))-- ( -28.80, z-score = -1.97, R) >droPer1.super_0 9481356 93 - 11822988 ---------AGAUUUUCAGACAA-----ACAUACAAACAGGGAGUACCGUAUUUCGCCGUAGAUAAGGGUGUACGGCGAGUGACACCAGUGG---GAGGCGGAACUCUGG-- ---------..............-----............(((((.((((..(((((((((.((....)).))))))))).....((....)---)..)))).)))))..-- ( -28.30, z-score = -2.12, R) >dp4.chr2 19808093 93 + 30794189 ---------UGAUUUUCAGACAA-----ACAUACAAACAGGGAGUACCGUAUUUCGCCGUAGAUAAGGGUGUACGGCGAGUGACACCAGUGG---GAGGCGGAACUCUGG-- ---------..............-----............(((((.((((..(((((((((.((....)).))))))))).....((....)---)..)))).)))))..-- ( -28.30, z-score = -1.97, R) >droAna3.scaffold_13340 4795367 95 + 23697760 ------------UUCCUCUCCUAAUUAUACUUACAUAAAGAGAAUACCUAAUCUCGCCAUAGAUUAAGGUAUACGGUGAGUGACACCAGUUG---GAUGUAGAGCUAUCC-- ------------...((((........(((((((.......(.(((((((((((......))))).)))))).).)))))))(((((....)---).)))))))......-- ( -20.10, z-score = -0.82, R) >droEre2.scaffold_4820 3167723 93 + 10470090 --------------UUAAGGCAAAUCAUACUCACAAACAGGGAAUAGCGGAUUUCGCCAUAAAUCAGUGUGUAUGGCGAGUGGCACCAAUUG---GAUGCGGAGCUCGGU-- --------------..........((((((.(((.....(((((........))).))........))).))))))(((((.(((((....)---).)))...)))))..-- ( -22.42, z-score = -0.23, R) >droYak2.chr3R 3084228 95 + 28832112 ------------UGGUGUGGCAAAUCAUACUCACAAACAGAGAAUAACGAAUUUCGCCAUAGAUCAGUGUGUAUGGCGAGUGGCACCAGUUG---GAGGCGGAGCUCUGU-- ------------((((((.((........(((.......)))...........((((((((.((....)).)))))))))).))))))...(---(((......))))..-- ( -30.10, z-score = -1.77, R) >droSec1.super_0 21094725 99 - 21120651 -----------UACUUAUGACAAAUCAUACUCACAAACAGAGAGUAGCGAAUUUCGCCAUAGAUCAGUGUGUAUGGCGAGUGGCACCAAUUGGAGGAGGCGGCGCUCGGU-- -----------(((((((((....)))))(((.......)))))))((.....((((((((.((....)).))))))))((.((.((.......))..)).)))).....-- ( -26.10, z-score = -0.64, R) >droSim1.chr3R 20582828 99 - 27517382 -----------UACUUAUGACAAAUCAUACUCACAAACAGAGAGUAGCGAAUUUCGCCAUAGAUCAGUGUGUAUGGCGAGUGGCACCAAUUGGAGGAGGCGGCGCUCGGU-- -----------(((((((((....)))))(((.......)))))))((.....((((((((.((....)).))))))))((.((.((.......))..)).)))).....-- ( -26.10, z-score = -0.64, R) >consensus ____________UCUUAUGACAAAUCAUACUCACAAACAGAGAGUAGCGAAUUUCGCCAUAGAUCAGUGUGUAUGGCGAGUGGCACCAAUUG___GAGGCGGAGCUCUGU__ ....................................................(((((((((.((....)).)))))))))................................ (-12.02 = -11.71 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:49 2011