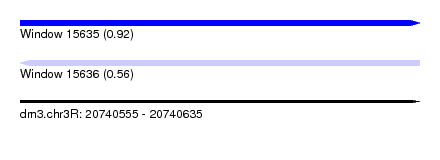
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,740,555 – 20,740,635 |
| Length | 80 |
| Max. P | 0.915654 |

| Location | 20,740,555 – 20,740,635 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 91.01 |
| Shannon entropy | 0.19434 |
| G+C content | 0.57902 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -29.23 |
| Energy contribution | -28.88 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.915654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
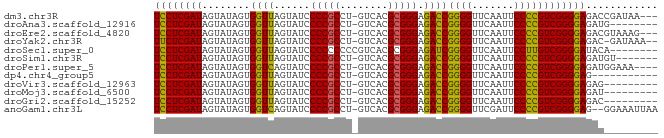
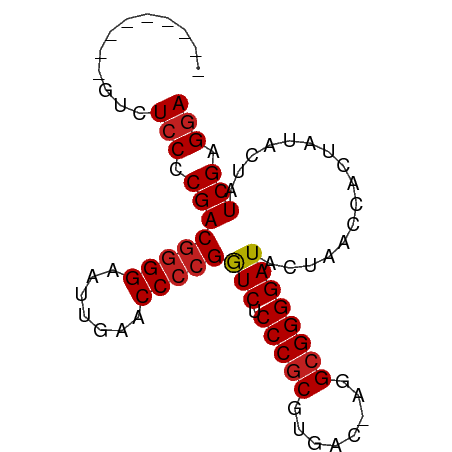
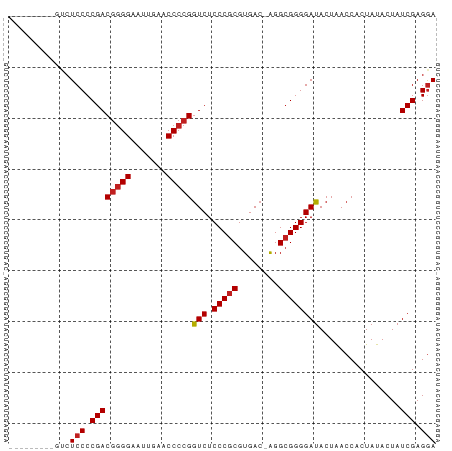
>dm3.chr3R 20740555 80 + 27905053 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACCGAUAA--- ((((((((........((((......(((((..-.....))))).))))((((.......)))))))))))).........--- ( -29.70, z-score = -0.80, R) >droAna3.scaffold_12916 5993929 75 - 16180835 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUG-------- ((((((((........((((......(((((..-.....))))).))))((((.......))))))))))))....-------- ( -29.70, z-score = -1.54, R) >droEre2.scaffold_4820 3161050 80 - 10470090 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGACGUAAAG--- ((((((((........((((......(((((..-.....))))).))))((((.......)))))))))))).........--- ( -29.70, z-score = -1.35, R) >droYak2.chr3R 3077535 80 - 28832112 UUCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAC-GAUAAA-- .((((((((.((.......)).))))(((.(((-(.....)))).(((.((((.......)))))))))))))).-......-- ( -27.90, z-score = -0.72, R) >droSec1.super_0 21088444 76 + 21120651 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCCCCCCGUCACGCGGGAGAUCGGGGUUCAAUUCCUUGUCGGGGAUACA-------- .((.(.........).))...((((((((..((((.....)))).(((.((((......)))).))))))))))).-------- ( -29.00, z-score = -1.49, R) >droSim1.chr3R 20575603 76 + 27517382 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGU------- ((((((((........((((......(((((..-.....))))).))))((((.......)))))))))))).....------- ( -29.70, z-score = -1.36, R) >droPer1.super_5 1386587 79 - 6813705 UCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUGGAAA---- ((((((((........((((......(((((..-.....))))).))))((((.......))))))))))))........---- ( -31.50, z-score = -1.31, R) >dp4.chr4_group5 1451821 72 - 2436548 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAG----------- ((((((((........((((......(((((..-.....))))).))))((((.......)))))))))))).----------- ( -29.70, z-score = -1.83, R) >droVir3.scaffold_12963 14518248 74 - 20206255 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAG--------- ((((((((........((((......(((((..-.....))))).))))((((.......))))))))))))...--------- ( -29.70, z-score = -1.65, R) >droMoj3.scaffold_6500 7169186 74 - 32352404 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAU--------- ((((((((........((((......(((((..-.....))))).))))((((.......))))))))))))...--------- ( -29.70, z-score = -1.51, R) >droGri2.scaffold_15252 7753518 74 + 17193109 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAC--------- ((((((((........((((......(((((..-.....))))).))))((((.......))))))))))))...--------- ( -29.70, z-score = -1.47, R) >anoGam1.chr3L 4285096 81 + 41284009 UCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCU-GUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGGAG--GGAAAUUAA ((((((((........((((......(((((..-.....))))).))))((((.......)))))))))))).--......... ( -31.50, z-score = -0.77, R) >consensus UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCU_GUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAC_________ ((((((((........((((......(((((........))))).))))((((.......))))))))))))............ (-29.23 = -28.88 + -0.36)
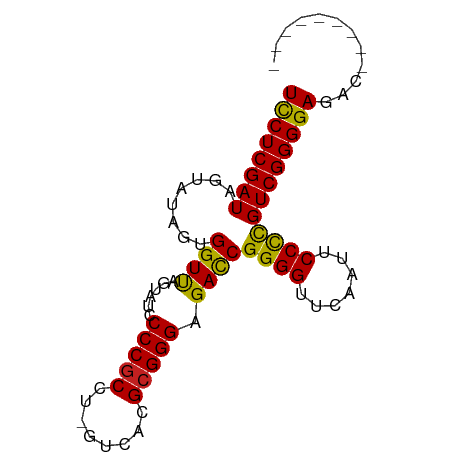
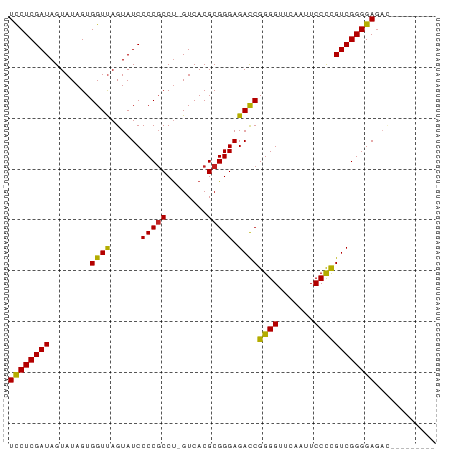
| Location | 20,740,555 – 20,740,635 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 91.01 |
| Shannon entropy | 0.19434 |
| G+C content | 0.57902 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -25.11 |
| Energy contribution | -25.37 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20740555 80 - 27905053 ---UUAUCGGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---.........(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -0.05, R) >droAna3.scaffold_12916 5993929 75 + 16180835 --------CAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA --------....(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -1.18, R) >droEre2.scaffold_4820 3161050 80 + 10470090 ---CUUUACGUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---.........(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -0.70, R) >droYak2.chr3R 3077535 80 + 28832112 --UUUAUC-GUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGAA --......-((.((((((.(((((.......)))))(((........)))-...)))))).))..................... ( -25.40, z-score = -0.64, R) >droSec1.super_0 21088444 76 - 21120651 --------UGUAUCCCCGACAAGGAAUUGAACCCCGAUCUCCCGCGUGACGGGGGGGGGAUACUAACCACUAUACUAUCGAGGA --------.((((((((..(((....)))..(((((.((........))))))).))))))))..................... ( -26.60, z-score = -0.87, R) >droSim1.chr3R 20575603 76 - 27517382 -------ACAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA -------.....(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -1.15, R) >droPer1.super_5 1386587 79 + 6813705 ----UUUCCAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUGACCACUAUACUAUCGAGGA ----........(((.(((.((((.......))))((((((((((.....-..)))))).....)))).........))).))) ( -27.60, z-score = -0.82, R) >dp4.chr4_group5 1451821 72 + 2436548 -----------CUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA -----------.(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -1.30, R) >droVir3.scaffold_12963 14518248 74 + 20206255 ---------CUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---------...(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -1.14, R) >droMoj3.scaffold_6500 7169186 74 + 32352404 ---------AUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---------...(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -1.10, R) >droGri2.scaffold_15252 7753518 74 - 17193109 ---------GUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---------...(((.((((((((.......)))))(((.(((((.....-..))))))))................))).))) ( -26.80, z-score = -0.88, R) >anoGam1.chr3L 4285096 81 - 41284009 UUAAUUUCC--CUCCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGAC-AGGCGGGGAUACUGACCACUAUACUAUCGAGGA ........(--(((......((((.......))))((((((((((.....-..)))))).....))))...........)))). ( -27.80, z-score = -0.57, R) >consensus _________GUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGAC_AGGCGGGGAUACUAACCACUAUACUAUCGAGGA ............(((.((((((((.......)))))(((.(((((........))))))))................))).))) (-25.11 = -25.37 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:46 2011