| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,705,883 – 20,705,997 |
| Length | 114 |
| Max. P | 0.562356 |

| Location | 20,705,883 – 20,705,997 |
|---|---|
| Length | 114 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.95 |
| Shannon entropy | 0.71198 |
| G+C content | 0.46869 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -10.65 |
| Energy contribution | -10.21 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
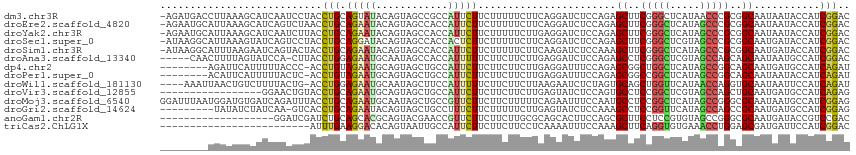
>dm3.chr3R 20705883 114 - 27905053 -AGAUGACCUUAAAGCAUCAAUCCUACCUGCAGUAUACAGUAGCCGCCAUUCUUCUUUUUCUUCAGGAUCUCCAGAGCUUCGGGCUCAUAACCCGCGGCAAUAAUACCAUCGGAC -.((((.(......)))))..(((...(((.......)))..(((((..................((....)).((((.....)))).......)))))............))). ( -24.50, z-score = -0.78, R) >droEre2.scaffold_4820 3122103 114 + 10470090 -AGAAUGCAUUAAAGCAUCAGUCUAACCUGCAGAAUACAGUAGCCACCAUUCUUCUUUUUCUUCAGGAUCUCCAGAGCUUCGGGCUCAUAGCCCGCGGCAAUAAUACCAUCGGAC -.((.(((......))))).((((..((((.((((...((.((........)).))..)))).)))).........(((.(((((.....))))).)))............)))) ( -29.20, z-score = -2.10, R) >droYak2.chr3R 3042003 114 + 28832112 -AGAAUGCAUUAAAGCAUCAAUCUUACCUGCAGAAUACAGUAGCCACCAUUCUUCUUUUUCUUGAGGAUCUCCAGAGCUUCGGGCUCAUAGCCCGCGGCAAUAAUACCAUCGGAC -.((.(((......)))))........((((........))))...((..(((((........)))))........(((.(((((.....))))).)))............)).. ( -26.70, z-score = -1.03, R) >droSec1.super_0 21053987 114 - 21120651 -AUAAGGCAUUAAAGUAUCAGUCCUACCUGCAGGAUACAGUAGCCACCACUCUUCUUUUUCUUCAGGAUCUCCAGAGCUUCGGGCUCGUAGCCCGCGGCAAUGAUACCAUCGGAC -.............((((((......((((.((((...((.((........)).))...)))))))).........(((.(((((.....))))).)))..))))))........ ( -29.00, z-score = -0.66, R) >droSim1.chr3R 20541017 114 - 27517382 -AUAAGGCAUUUAAGAAUCAGUACUACCUGCAGAAUACAGUAGCCACCAUUCUUCUUUUUCUUCAAGAUCUCCAAAGCUUCGGGCUCAUAGCCCGCGGCAAUGAUACCAUCGGAC -..((((.....((((((.....((((.((.......)))))).....)))))).....)))).......(((...(((.(((((.....))))).))).(((....))).))). ( -22.80, z-score = -0.45, R) >droAna3.scaffold_13340 4754408 109 + 23697760 -----CAACUUUUAGUAUCCA-CUUACCUGGAGAAUGCAAUAGCCACCAUUUUUCUUCUUCUUGAGGAUCUCCAGAGCCUCGGGCUCGUAGCCAGCAGCAAUAAUGCCAUCGGAC -----............(((.-.....(((((((..((....)).........(((((.....)))))))))))).((....(((.....)))....))............))). ( -26.40, z-score = -0.80, R) >dp4.chr2 20747655 106 + 30794189 --------AGAUUCAUUUUUACCC-ACCUGUAGAAUGCAGUAGCUGCCAUUCUUCUUCUUCUUGAGGAUUUCCAGAGCGGCUGGCUCAUAGCCGGCAGCAAUGAUGCCAUCAGAU --------.((((((((.......-..((((.....))))..((((((.(((((((((.....))))).....)))).(((((.....))))))))))))))))....))).... ( -30.10, z-score = -0.82, R) >droPer1.super_0 10435729 106 - 11822988 --------ACAUUCAUUUUUACUC-ACCUGUAGAAUGCAGUAGCUGCCAUUCUUCUUCUUCUUGAGGAUUUCCAGAGCGGCCGGCUCAUAGCCGGCAGCAAUAAUACCAUCAGAU --------................-..(((.(((((((((...))).))))))(((((.....)))))....))).((.((((((.....)))))).))................ ( -29.40, z-score = -2.26, R) >droWil1.scaffold_181130 3942952 110 + 16660200 ----AAAUUAACUGUCUUUUACUG-ACCUGGAGAAUGCAAUAGCUUCCAUUUUUCUUCUUCUUAAGAAUCUCUAGUGCAGCUGGUUCAUAACCAGGUGCAAUAAUUCCAUCAGAU ----.................(((-(.(((((((..((....))........(((((......))))))))))))((((.(((((.....))))).)))).........)))).. ( -28.10, z-score = -2.78, R) >droVir3.scaffold_12855 2436855 98 - 10161210 -----------------GGAACUGUACCUGCAGAAUGCAGUAGCUGCCAUUCUUCUUCUUCUUGAGUAUCUCCAGUGCCUCCGGCUCGUAGCCAGCUGCAAUGAUGCCAUCAGAG -----------------....(((.....(((...((((((.(((((.((((...........)))).......(.(((...))).))))))..))))))....)))...))).. ( -24.00, z-score = 0.67, R) >droMoj3.scaffold_6540 11647301 115 - 34148556 GGAUUUAAUGGAUGUGAUCAGAUUUACCUGCAGAAUGCAAUAGCUGCCGUUCUUCUUCUUUUUCAGAAUUUCCAAUGCCUCCGGCUCAUAGCCGGGCGCAAUAAUGCCAUCGGAG (((..(..((((...((.(((......))).((((((((.....)).))))))...))...))))..)..)))..(((.((((((.....)))))).)))............... ( -27.50, z-score = 0.48, R) >droGri2.scaffold_14624 1729231 105 + 4233967 ---------UAUAUCUAUCAA-GUCACCUGCAGAAUACAGUAGCUGCCUUUCUUCUUUUUCUUGAGUAUCUCCAAAGCCUCCGGUUCAUAGCCAGCCGCAAUGAUGCCAUCGGAG ---------........((((-(......(.((((..(((...)))...)))).).....))))).............(((((((((((.((.....)).))))....))))))) ( -18.90, z-score = 0.15, R) >anoGam1.chr2R 57978899 96 + 62725911 -------------------GGAUCGAUCUGCAGCACGCAGUACGAACCGUUCUUCUUCUUGCGCAGCACUUCCAGCGCUUCCUCCGUGUAGCCGGGCGCAAUGAUACCGUCCGAC -------------------((.(((..((((.....))))..))).))(..(....(((((((((((.(.....).)))....(((......))))))))).))....)..)... ( -25.70, z-score = 0.56, R) >triCas2.ChLG1X 563097 90 + 8109244 -------------------------AUUUGAAGGACACAGUAAUUGCCAUUCUUCUUCUUCCUCAAAAUUUCCAAAGCUUCAGGUGUGAAACCUGGAGCGAUGAUUCCAUCGGAC -------------------------.(((((.(((...((..............))...))))))))...(((...(((((((((.....)))))))))((((....))))))). ( -26.74, z-score = -2.62, R) >consensus ________AUUAAAGUAUCAAUCCUACCUGCAGAAUACAGUAGCUGCCAUUCUUCUUCUUCUUCAGGAUCUCCAGAGCUUCGGGCUCAUAGCCCGCGGCAAUAAUACCAUCGGAC ...........................(((.(((((............))))).......................((..(((((.....)))))..))...........))).. (-10.65 = -10.21 + -0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:41 2011