| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,703,340 – 20,703,457 |
| Length | 117 |
| Max. P | 0.602311 |

| Location | 20,703,340 – 20,703,457 |
|---|---|
| Length | 117 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.35 |
| Shannon entropy | 0.73988 |
| G+C content | 0.51619 |
| Mean single sequence MFE | -35.26 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.36 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20703340 117 + 27905053 CUGCUCGCCAAGGACACUUCUCCGAUUGUGGUGGGCGGCACGAAUUACUACAUAGAAUCCCUACUUUGGGAUAUUCUGGUUGACUCGGAUGUCAAGCCGGACGAAGGCAAACAUUCG--- ((((((((((.(((......))).....))))))))))..(((((...........(((((......)))))..((((((((((......))).)))))))...........)))))--- ( -40.00, z-score = -2.65, R) >apiMel3.Group11 7506515 120 + 12576330 CUUCUCGCGAGGAGGAAGCUCCCGAUCAUCGUUGGUGGCACCAACUAUUACAUAGAGUCCAUCCUUUGGGAAGUUCUGAUGAACGACUCGAGGAUCGGCAAGGACGAUGAUGACCGGUUG ....(((((.((((....))))))(((((((((((.....)))))...........((((((((((.(((..((((....))))..)))))))))......))))))))))...)))... ( -40.30, z-score = -1.03, R) >droEre2.scaffold_4820 3119571 117 - 10470090 CUGCUCGCCAAGGACUCUCCACCGAUUGUGGUGGGCGGCACCAAUUACUACAUAGAAUCCCUGCUUUGGGAUGUUCUGGUGGAUUCGGAUGUCAAGCUGGACAAAGGCAAACCUUCG--- ......(((..((.....)).((((...(((((.....)))))....((((.(((((((((......)))))..))))))))..)))).((((......))))..))).........--- ( -35.60, z-score = -0.28, R) >droYak2.chr3R 3039456 117 - 28832112 CUGCUCGCCAAGGAAACUCCUUCGAUUAUUGUGGGCGGCACCAAUUACUACAUAGAAUCCCUGCUUUGGGAUGUUCUGGUGGACUCGGAUGUCAAGCCGGACGAAUGCAAACCUCCG--- .((((((((((((.....))))...........(((((((((.....((((.(((((((((......)))))..))))))))....)).))))..))))).)))..)))........--- ( -32.90, z-score = -0.34, R) >droSec1.super_0 21051430 117 + 21120651 CUGCUCGCCAAGGAAACUCCUCCGAUUGUAGUGGGCGGCACGAAUUACUACAUAGAAUCCCUGCUUUGGGAUAUUCUGGUGGACUCGGAUGUCAAGCCGGACGAACACAAAACUUCG--- ((((((((((.(((......)))...))..))))))))..((((............(((((......)))))..((((((.(((......)))..))))))............))))--- ( -32.20, z-score = -0.53, R) >droSim1.chr3R 20538462 117 + 27517382 CUGCUCGCCAAGGACACUCCUCCGAUUGUGGUGGGCGGCACGAAUUACUACAUAGAAUCCCUGCUUUGGGAUAUUCUGGUGGACUCGGAUGUCAAGCCGGACGAACACAAACCUUCG--- ((((((((((.(((......))).....))))))))))..((((............(((((......)))))..((((((.(((......)))..))))))............))))--- ( -37.80, z-score = -1.55, R) >droAna3.scaffold_13340 4751793 111 - 23697760 CUGCUCGCUAAAGCCAAGCCGCCCAUAGUGGUCGGUGGAACCAAUUACUACAUCGAGUCAUUGUUAUGGGACAUUCUCGUGGACUCUCAAGAGAAGCCCGCUGCCCUCUUG--------- ..(((......)))...((.((..(((((((((((((.............)))))).)))))))...(((...(((((.(((....))).))))).))))).)).......--------- ( -28.22, z-score = 0.44, R) >dp4.chr2 20745155 117 - 30794189 CUGCUGGCCAAAUCCACGCCGCCCAUUGUCGUGGGCGGCACCAAUUAUUACAUUGAGUCCCUGCUGUGGGACAUAUUGGUGAACACG--CAGGAUGGGGAUAGCAACAAGAGCCCGACG- .(((((.((..((((..(((((((((....)))))))))((((((((......)))(((((......)))))...))))).......--..))))..)).)))))..............- ( -47.50, z-score = -2.72, R) >droPer1.super_0 10433223 117 + 11822988 CUGCUGGCCAAAUCCACGCCGCCCAUUGUCGUGGGUGGCACCAAUUAUUACAUUGAGUCCCUGCUUUGGGACAUAUUGGUGAGCACG--CAGGAUGGGGAUAGCAACAAGAGCCCGACG- .(((((.((..((((..(((((((((....)))))))))((((((((......)))(((((......)))))...))))).......--..))))..)).)))))..............- ( -45.60, z-score = -2.19, R) >droWil1.scaffold_181130 3940451 114 - 16660200 CUGCUGGCCAAAUCCCAAUCACCCAUUAUUGUUGGUGGCACCAAUUACUAUAUUGAGUCACUGUUGUGGGACAUUCUGGUGAAUACCAGCGGCGACGAGGACCUGGAUGAAACC------ .......(((..(((......(((((......(((((((..((((......)))).)))))))..))))).....(((((....)))))((....)).)))..)))........------ ( -27.60, z-score = 0.96, R) >droVir3.scaffold_12855 2433794 111 + 10161210 CUGCUGGCCAAAUCGAAGCCGCCAAUUGUGGUCGGCGGCACCAACUACUACAUCGAAUCCUUGCUGUGGGACAUAUUGGUUAGCAC---CAAAGAGGAGCAGGCAGCAACAGUU------ .(((((.((.((((((.((((((.((....)).))))))........(((((.(((....))).)))))......)))))).((.(---(.....)).)).)))))))......------ ( -36.10, z-score = -0.81, R) >droMoj3.scaffold_6540 11644609 111 + 34148556 CUGCUGGCUAAAUCAAAACCACCGAUUGUGGUUGGCGGUACAAAUUAUUAUAUAGAAUCUUUACUGUGGGACAUACUGGUGGAUAC---CAAAGAGGAGAAGGUAACUACAAAC------ ((((..((((((((.........)))).))))..))))...................((((..((.(((..(((....)))....)---)).))..))))..............------ ( -22.80, z-score = -0.13, R) >droGri2.scaffold_14624 1726553 108 - 4233967 CUGUUGGCCAAAUCGAAGCCACCAAUUGUUGUUGGCGGCACCAAUUACUACAUCGAAUCCCUGCUCUGGGAUAUACUGGUAAACAC---CAAGGAUGAGCAGACCGCAGAU--------- ((((.((.(........(((.(((((....))))).)))...........((((..(((((......)))))....((((....))---))..))))....).))))))..--------- ( -31.70, z-score = -1.54, R) >consensus CUGCUCGCCAAAGCCACGCCACCGAUUGUGGUGGGCGGCACCAAUUACUACAUAGAAUCCCUGCUUUGGGACAUUCUGGUGGACACG_ACGUCAAGGCGGACGAAGCCAAACCU__G___ ((((((((......................)))))))).........((((.....(((((......)))))......))))...................................... (-13.48 = -13.36 + -0.12)

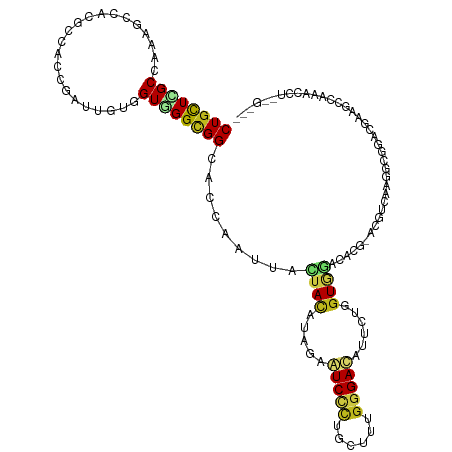
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:40 2011