| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,632,026 – 20,632,132 |
| Length | 106 |
| Max. P | 0.990906 |

| Location | 20,632,026 – 20,632,132 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 66.27 |
| Shannon entropy | 0.70743 |
| G+C content | 0.50872 |
| Mean single sequence MFE | -37.91 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.67 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.990906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
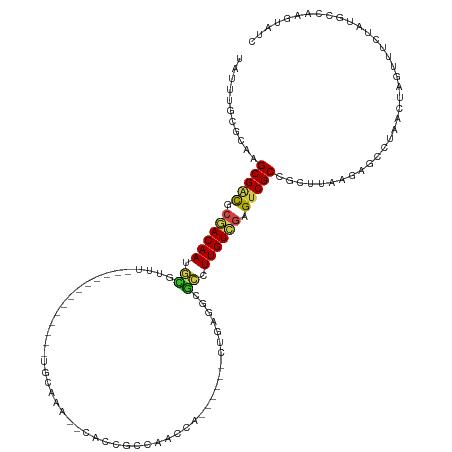
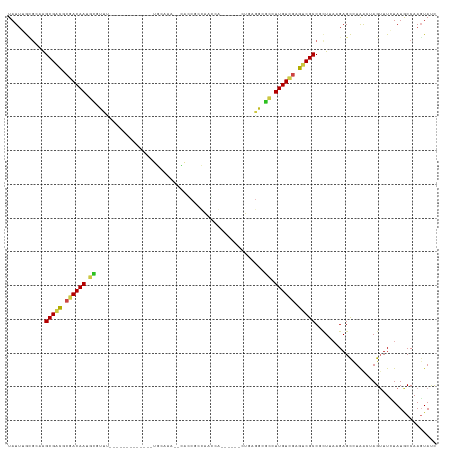
>dm3.chr3R 20632026 106 - 27905053 UAUUGGCGCAAGCGACACGACAAUGCGUUU-------------UGUAUA--AACCGACAGGAG------GUGAGGCGCCUUGUCGAGUCGCCGCUAAAGAGCCUAACAAGUUUCUAUACCAAGUAUC ..((((((...(((((.((((((.((((((-------------..(...--..((....))..------)..)))))).)))))).)))))))))))((((.((....)).))))............ ( -36.10, z-score = -2.97, R) >droGri2.scaffold_14906 5105381 124 - 14172833 UAUUUAAUACGGCGUCGCGACAAACUCUUGCA-AACACUACAAAGUCAGCGUUGCGCUGACAAACUG--CUAACACAUUUUGUCGAGUCGCCCCUUAAGAGUCUAGCUAGUUUAUAUGCCAAGUACU ..........(((((.((((.......)))).-...........(((((((...)))))))((((((--((((((.....))).((.((.........)).)))))).)))))..)))))....... ( -28.30, z-score = -0.55, R) >droMoj3.scaffold_6540 28951337 121 + 34148556 UAUUUAAGACGGCGGCGCAACAAGAGCUCGCA----CUGGCAGCGCCAGCGCUACGCUGAUCAGCUG--CUCAGGCGCUUUGUUGAGUCGCCCCUGAAGAGUCUAACUAGUUUAUAUGCCAAUUACU ......((((((((((.(((((.((((..((.----((((((((..(((((...)))))....))))--).)))))))))))))).)))))).(....).))))....................... ( -44.40, z-score = -1.63, R) >droVir3.scaffold_13047 18055605 121 + 19223366 UAUUUAAGACAGCGACGCGACAAAGCCUUGC----CAUGCCAGCGCCAACGUUACGCUGACCAGCUG--CUCAGGCGCUUUGUCGAGUCGCCCUUUACGAGUCUAACUAGUUUAUAUGCCAAGUACU ......((((.(((((.(((((((((((((.----...(((((((.........)))))....))..--..)))).))))))))).))))).(.....).))))....................... ( -36.80, z-score = -1.82, R) >droWil1.scaffold_181130 9182581 127 - 16660200 UAUAUGUAUGAGCGAUGCGACAAAGUGUCAAGGAGUCAGCAGCAGCAAUUGUUGCAGCAAAUGAUUGUUAUAAUUCGCUUUGUCGAGUCGCCCCUUAAGAGCUUAAGUAGUUUUUAUGCCAAGUAUC .....(((((((((((.((((((((((...((.(((((((.(((((....))))).))...))))).))......)))))))))).)))))..(((((....)))))......))))))........ ( -38.70, z-score = -3.44, R) >droPer1.super_7 2698353 118 + 4445127 UAUUUGCGGAAGCGACGGCACAACGUGUCU---CUCCGGCAGCUGCAGAGUCACCUCCAGCCG------CUGAGACGCUUUGUCGAGUCGCCGCUUACGAGCUUAAGUAGAUUCUAUGCCAAGUACC ...((((....)))).(((((((.((((((---(...(((.((((.((......)).)))).)------))))))))).)))).(((((...(((((......))))).)))))...)))....... ( -42.40, z-score = -1.81, R) >dp4.chr2 29198200 118 + 30794189 UAUUUGCGGAAGCGACGGCACAACGUGUCU---CUCCGGCAGCUGCAGAGUCACCUCCAGCCG------CUGAGACGCUUUGUCGAGUCGCCGCUUACGAGCUUAAGUAGAUUCUAUGCCAAGUACC ...((((....)))).(((((((.((((((---(...(((.((((.((......)).)))).)------))))))))).)))).(((((...(((((......))))).)))))...)))....... ( -42.40, z-score = -1.81, R) >droAna3.scaffold_13340 3303089 121 + 23697760 UAUUUGUGCAAGCGUCGCGACAACGCGUC----UCUCGUCAGGCCCAAUCAGUGCGCCCACCACCUG--ACCAGACGCCUUGUCGAGUCGCCGCUGCAGAGCCUGACUAGUUUAUAUGCCAAUUAUC ...(((.(((.(((.(.((((((.(((((----(...((((((........(((....)))..))))--)).)))))).)))))).).))).(((....)))..............))))))..... ( -37.60, z-score = -1.96, R) >droEre2.scaffold_4820 3045012 106 + 10470090 UAUUGGCGCAAGCGACACGACAAUGCGUUU-------------UGUAUA--CACCGCCACGCG------GUGAGGCGCCUUGUCGAGUCGCCGCUCAAGAGCCUAACUAGUUUCUAUACCAAGUAUC ..((((.((..(((((.((((((.((((((-------------......--((((((...)))------))))))))).)))))).))))).))...((((.((....)).))))...))))..... ( -38.40, z-score = -2.88, R) >droYak2.chr3R 2966539 106 + 28832112 UAUUGGCGCAAGCGACACGACAAUGCGUUU-------------UGUAUA--CACCGAGACGCG------GUGAGGCGCCUUGUCGAGUCGCCGCUAAAGAGCCUAACUAGUUUCUAUACCAAGUAUC ..((((((...(((((.((((((.((((((-------------......--(((((.....))------))))))))).)))))).)))))))))))((((.((....)).))))............ ( -37.80, z-score = -2.85, R) >droSec1.super_0 20967612 106 - 21120651 UAUUGGCGCAAGCGACACGACAAUCCGUUU-------------UGCAUA--CACCGACAGGCA------GUGAGGCGGCUUGUCGAGUCGCCGCUAAAGAGCCUAACAAGUUUCUAUACCAAGUAUC ..((((((...(((((.((((((.((((((-------------..(...--..((....))..------)..)))))).)))))).)))))))))))((((.((....)).))))............ ( -37.00, z-score = -3.30, R) >droSim1.chr3R 20467509 106 - 27517382 UAUUGGCGCAAGCGACACGACAAUGCGUUU-------------UGUAUA--CACCGACAGGCA------GUGAGGCGCCUUGUCGAGUCGCCGCUAAAGAGCCUAACAAGUUUCUAUACCAAGUAUC ..((((((...(((((.((((((.((((((-------------..(...--..((....))..------)..)))))).)))))).)))))))))))((((.((....)).))))............ ( -35.00, z-score = -2.57, R) >consensus UAUUUGCGCAAGCGACGCGACAAUGCGUUU_____________UGCAAA__CACCGCCAACCA______CUGAGGCGCCUUGUCGAGUCGCCGCUUAAGAGCCUAACUAGUUUCUAUGCCAAGUAUC ...........(((((.((((((.((..................................................)).)))))).))))).................................... (-13.57 = -13.67 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:33 2011