
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,623,109 – 20,623,264 |
| Length | 155 |
| Max. P | 0.714691 |

| Location | 20,623,109 – 20,623,264 |
|---|---|
| Length | 155 |
| Sequences | 6 |
| Columns | 181 |
| Reading direction | forward |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.63061 |
| G+C content | 0.38788 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -16.24 |
| Energy contribution | -19.30 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20623109 155 + 27905053 AGAGGGGCACGAAGGCUGUUUUUCAGUAAUAUUGGGGUCAGCAUCUCCUGAAAAU-----------UACAUACAAUAUGGUUUCAAAAUUGUAAUUCUGUUUUUAUUAUAA----------AGUUGUUCGACAUUUUUAUGCAACGUGCCCCUCCAUAGACUACCAUAU---AUCGAGC-- .((((((((((...((((..(..((((...))))..).))))......(((((((-----------....((((((...........)))))).....))))))).(((((----------((.((.....)).)))))))...))))))))))...............---.......-- ( -38.00, z-score = -1.74, R) >droEre2.scaffold_4820 3036064 174 - 10470090 AGAGGGGCACGUAUGCUGUUCGUCAGUAAUGUCGGGGUCACCAUCUCCUGAAAAUGG-CAGAGUCACACAUGCAAGAUGGUUUCAUCCAUUUAAUUCUGUUUU-AUUAUAACGUUAUACACAGUUGUUCAACAUUUUCAUUCACCGUGCCCCUCUCUAGACUACCAUAU---ACAAAGU-- (((((((((((..(((((.....))))).....((((.......))))(((((((((-((((((..........((((((......)))))).)))))))...-...(((((..........)))))....)))))))).....)))))))))))..............---.......-- ( -47.70, z-score = -2.82, R) >droYak2.chr3R 2957607 175 - 28832112 AGAGGGGCACGAGAGCUGUUUGUCGUUACCAUCGGGGUCACCAUCUCCUAAAAAACGGCGGAGUCACACAUGCAAGAUGGUUUCAUCAUUUUAAUUCUGUUUA-AUUAUCAAGUUAUAUAAAGUUGUUCAACGUUUUUAUUCACCGUGCCCCUCUCUAGACUAUCAUAC---AUCCAGU-- (((((((((((.((((....((..(..((((((((.....))..((((...........))))............))))))..)..))....((((.(((.((-(((....))))).))).))))))))...(........)..)))))))))))..............---.......-- ( -42.40, z-score = -1.61, R) >droSec1.super_0 20958665 154 + 21120651 AGAGGGGUACGAAGGCAGUUUUUCAGUAAAAUCGGGGUCACCAUCUCCUGAAAAU-----------UACAUACAAUAUGGUUUCAAAAUUGUAGUUCUGUUUU-AUUAUAA----------AGUUCUUCGUCAUUUUUAUGCACCGAGCCACUCUCUAGACUAACAUAU---AUCCAGU-- (((((((((..(((((((..((.((((......((((.......))))(((((..-----------..((((...)))).)))))..)))).))..)))))))-..)))..----------.((((...(((((....))).)).))))..))))))............---.......-- ( -26.90, z-score = 0.51, R) >droSim1.chr3R 20458064 153 + 27517382 GGAGGGGUACGAAGGCUGUUUUUCAGUAAUAUCGGGGUCACCAUCUCCUGAAAAU-----------UACAUACAAUAUGGUUUCAAAAUUGCAAUUCUGUUUU-AUUAUAA----------AGUUCUUCGACAUUUUUAUGCACCGUGCCCCUCUCUAGAC-AACAUAU---AUCGAGU-- (((((((((((((((((........(((((.((((((........))))))..))-----------)))(((.((((..(((.((....)).)))..)))).)-)).....----------)).)))))(.(((....))))...))))))))))......-.......---.......-- ( -34.90, z-score = -1.25, R) >droMoj3.scaffold_6540 28939681 143 - 34148556 --------------GCUACAUUACAGUACAACUCGGUUUGG---UUAAUGAAUGG-----------AUUAUACAAAGUUAGU--AACGGUGCAAUUUAAUAAAAGCUAGUGU--------UGGCAGACAAGUGAUGAUCAGAGAUGUGACAAUCGCUAGACCACCACAAGCAAUCGAGAUC --------------.................(((((((...---........(((-----------(((.(((...((....--.)).))).))))))......(((.(((.--------(((....(.((((((..(((......)))..)))))).).))).))).))))))))))... ( -28.70, z-score = 0.22, R) >consensus AGAGGGGCACGAAGGCUGUUUUUCAGUAAUAUCGGGGUCACCAUCUCCUGAAAAU___________UACAUACAAUAUGGUUUCAAAAUUGUAAUUCUGUUUU_AUUAUAA__________AGUUGUUCAACAUUUUUAUGCACCGUGCCCCUCUCUAGACUACCAUAU___AUCGAGU__ .((((((((((...((((.....))))....((((((........)))))).............................................................................................))))))))))........................... (-16.24 = -19.30 + 3.06)
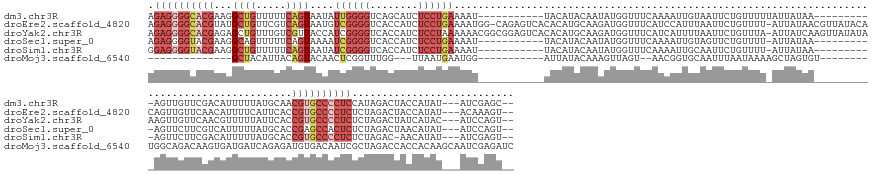

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:33 2011