
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,542,951 – 20,543,053 |
| Length | 102 |
| Max. P | 0.714417 |

| Location | 20,542,951 – 20,543,053 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.03 |
| Shannon entropy | 0.73192 |
| G+C content | 0.46799 |
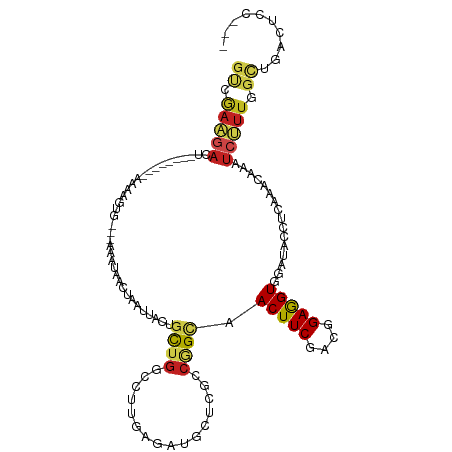
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -7.78 |
| Energy contribution | -8.12 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714417 |
| Prediction | RNA |
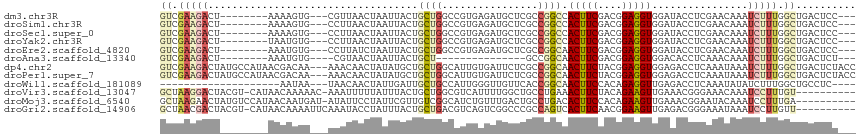
Download alignment: ClustalW | MAF
>dm3.chr3R 20542951 102 - 27905053 GUCGAAGACU--------AAAAGUG---CGUUAACUAAUUACUGCUGGCCGUGAGAUGCUCGCCGGCCACUUCGACGGAGGUGGAUACCUCGAACAAAUCUUUGGCUGACUCC--- (((((((...--------......(---((.(((....))).)))((((((((((...)))).))))))))))))).(((((....)))))......................--- ( -31.90, z-score = -1.96, R) >droSim1.chr3R 20378214 102 - 27517382 GUCGAAGACU--------AAAAGUG---CCUUAACUAAUUACUGCUGGCCGUGAGAUGCUCGCCGGCCACUUCGACGGAGGUGGAUACCUCGAACAAAUCUUUGGCUGACUCC--- (((((((...--------...(((.---.....))).........((((((((((...)))).))))))))))))).(((((....)))))......................--- ( -31.70, z-score = -2.26, R) >droSec1.super_0 20881152 102 - 21120651 GUCGAAGACU--------AAAAGUG---CCUUAACUAAUUACUGCUGGCCGUGAGAUGCUCGCCGGCCACUUCGACGGAGGUGGAUACCUCGAACAAAUCUUUGGCUGACUCC--- (((((((...--------...(((.---.....))).........((((((((((...)))).))))))))))))).(((((....)))))......................--- ( -31.70, z-score = -2.26, R) >droYak2.chr3R 2877779 102 + 28832112 GUCGAAGACU--------UAAUGUG---CCUUAACUAAUUACUGCUGGCCGUGAGAUGCUCGCCGGCCACUUCGACGGAGGUGGAUACCUCGAACAAAUCUUUGGCUGACUCC--- (((((((...--------......(---(..(((....)))..))((((((((((...)))).))))))))))))).(((((....)))))......................--- ( -30.40, z-score = -1.96, R) >droEre2.scaffold_4820 2956698 102 + 10470090 GUCGAAGACU--------AAAUGUG---CCUUAUCUAAUUACUGCUGGCCGUGAGAUGCUCGCCGGCAACUUCGACGGAGGUGGAUACCUCGAACAAAUCUUUGGCUGACUCC--- (((((((((.--------....)).---..............(((((((.((.....))..))))))).))))))).(((((....)))))......................--- ( -28.40, z-score = -1.32, R) >droAna3.scaffold_13340 19065371 86 + 23697760 GUCGAAGACU--------AAAUGUG----CGUAACUAAUUACUGCU---------------GCCGGCAACUUCGACGGAGGUGGACACCUCAAACAAAUCUUUGGCUGACUCU--- (((((((...--------.......----.((((....))))(((.---------------....))).))))))).(((((....)))))......................--- ( -20.90, z-score = -1.35, R) >dp4.chr2 29104275 113 + 30794189 GUCGAAGACUAUGCCAUAACGACAA---AAACAACUAUAUGCUGCUGGCAUUGUGAUUCUCGCCGGCAACUUCUACGGAGGUGGAGACCUCAAAUAAAUCUUUGGCUGACUCUACC ((((((((.................---..............(((((((............))))))).........(((((....))))).......)))))))).......... ( -26.70, z-score = -1.45, R) >droPer1.super_7 2601337 113 + 4445127 GUCGAAGACUAUGCCAUAACGACAA---AAACAACUAUAUGCUGCUGGCAUUGUGAUUCUCGCCGGCAACUUCUACGGAGGUGGAGACCUCAAAUAAAUCUUUGGCUGACUCUACC ((((((((.................---..............(((((((............))))))).........(((((....))))).......)))))))).......... ( -26.70, z-score = -1.45, R) >droWil1.scaffold_181089 2822250 89 + 12369635 --------------------AAUAA---UAACAACUAUUGAUUGCUGCCAUUGGGUUGUUCACCGGCAACUUCCACAGAGGUUGAGACCUCAAAUAUAUCUUUGGCUGCCUC---- --------------------.....---...............((.((((..(((((((......))))))......(((((....)))))........)..)))).))...---- ( -21.10, z-score = -1.09, R) >droVir3.scaffold_13047 850008 104 - 19223366 GCUAAGGACUACGU-CAUAACAAAAAC-AAAUUUUUAUUUACUGCUGGCGUCAUUUUGGCUGCCUGAAACUUCUACAGAAGUUGAAACGGGAAACAAAUCCUUUGU---------- ((.(((((..((((-((..........-.................))))))...((((....((((.((((((....))))))....))))...))))))))).))---------- ( -20.73, z-score = -1.06, R) >droMoj3.scaffold_6540 4844598 105 - 34148556 GCUAAGAACUAUGUCCAUAACAAUGAU-AUAUUCCUAUUCGUUGUCGGCAUCUGUUUGACUGCCUGACACUUCCACAGAAGUUGAAACGGAAUACAAAUCCUUUGA---------- ....................(((.(((-.((((((..((((..(((((((((.....)).)))).)))(((((....)))))))))..))))))...)))..))).---------- ( -21.30, z-score = -1.37, R) >droGri2.scaffold_14906 8716896 105 + 14172833 GCUAACGACUACGU-CAUAACAAAAUUCAAAUACCUAUUUACUGCUGACGUCAGUCGGCCCGCCAGUCACUUCAACGGAAGUUGAGACGGGAAAUAAAUCCUUGUU---------- ((...(((((((((-((.....((((..........)))).....)))))).)))))....))..(((...(((((....))))))))((((......))))....---------- ( -26.40, z-score = -2.25, R) >consensus GUCGAAGACU________AAAAGUG___AAAUAACUAAUUACUGCUGGCCUUGAGAUGCUCGCCGGCAACUUCGACGGAGGUGGAUACCUCAAACAAAUCUUUGGCUGACUCC___ ((.(((((...................................((((................)))).(((((....)))))................))))).)).......... ( -7.78 = -8.12 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:28 2011