| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,518,222 – 20,518,313 |
| Length | 91 |
| Max. P | 0.997600 |

| Location | 20,518,222 – 20,518,313 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 60.65 |
| Shannon entropy | 0.73007 |
| G+C content | 0.31589 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -5.20 |
| Energy contribution | -5.59 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.997600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
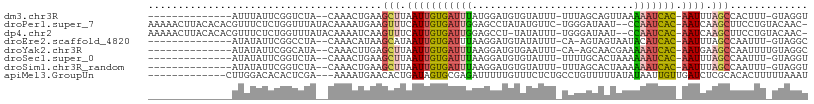
>dm3.chr3R 20518222 91 + 27905053 --------------AUUUAUUCGGUCUA--CAAACUGAAGCUUAAUUGUGAUUUAUGGAUGUGUAUUU-UUUAGCAGUUAAAAAUCAC-AAUUUAGCCACUUU-GUAGGU --------------..........((((--((((.((..(((.(((((((((((.(((.(((......-....))).))).)))))))-)))).))))).)))-))))). ( -24.20, z-score = -3.53, R) >droPer1.super_7 2576082 105 - 4445127 AAAAACUUACACACGUUUCUCUGGUUUAUACAAAAUGAAGUUUCAUUGUGAUUGGAGCCUAUAUGUUC-UGGGAUAAU--CCAAUCAC-AAUCAAGCUUCCUGUACAAC- ..(((((...............))))).((((....(((((((.((((((((((((.((((.......-))))....)--))))))))-))).))))))).))))....- ( -29.66, z-score = -4.26, R) >dp4.chr2 29079681 104 - 30794189 AAAAACUUACACACGUUUCUCUGGUUUAUACAAAAUCAAGUUUCAUUGUGAUUGGAGCCU-UAUAUUU-UGGGAUAAU--CCAAUCAC-AAUCAAGCUUCCUGUACAAC- .......((((...........(((((.....)))))((((((.((((((((((((.(((-.......-.)))....)--))))))))-))).))))))..))))....- ( -27.20, z-score = -4.42, R) >droEre2.scaffold_4820 2930076 90 - 10470090 --------------AUAUAUUCGGCCUA--CAAACAUAAGCAUAAUUGUGAUUUAAGGAUGUAUAUUU-CA-AGUAGUAAUACAUCAC-AAUUUAGCCAAUUU-GUAGGC --------------.........(((((--((((.....((..(((((((((....(((.......))-).-.(((....))))))))-))))..))...)))-)))))) ( -23.80, z-score = -4.18, R) >droYak2.chr3R 2853572 91 - 28832112 --------------AUAUAUUCGGCAUA--CAAACUUGAGCUUAAUUGUGAUUUAAGGAUGUGAAUUU-CA-AGCAACGAAAAAUCAC-AAUGAAGCCAAUUUUGUAGGC --------------.........((.((--((((.(((.((((.((((((((((..(..(((......-..-.))).)...)))))))-))).))))))).)))))).)) ( -24.20, z-score = -3.50, R) >droSec1.super_0 20852382 91 + 21120651 --------------AUAUAUUCGGUCUA--CAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUU-UUUUGCACUAAAAAAUCAC-AAUUUAGCCAAUUU-GUAGGU --------------..........((((--((((.((..(((.(((((((((((......(((((...-...)))))....)))))))-)))).))))).)))-))))). ( -27.20, z-score = -4.90, R) >droSim1.chr3R_random 984822 91 + 1307089 --------------AUAUAUUCGGUCUA--CAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUU-UUUAGCACUAAAAAAUCAC-AAUUUAGCCAAUUU-GUAGGU --------------..........((((--((((.((..(((.(((((((((((......((((....-....))))....)))))))-)))).))))).)))-))))). ( -25.40, z-score = -4.30, R) >apiMel3.GroupUn 298547683 94 - 399230636 -------------CUUGGACACACUCGA---AAAAUGAACACUGAUAGUGCGAGAUUUUUGUUUCUCUGCCUGUUUUUAUAUAAUUGUUGAUCUCGCACACUUUUUAAAU -------------...............---................((((((((((...(((...................)))....))))))))))........... ( -14.51, z-score = -0.31, R) >consensus ______________AUAUAUUCGGUCUA__CAAACUGAAGCUUAAUUGUGAUUUAAGGAUGUGUAUUU_UUUAGCAAUAAAAAAUCAC_AAUUUAGCCAAUUU_GUAGGU .......................................(((.(((((((((((...........................))))))).)))).)))............. ( -5.20 = -5.59 + 0.39)
| Location | 20,518,222 – 20,518,313 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 60.65 |
| Shannon entropy | 0.73007 |
| G+C content | 0.31589 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -2.83 |
| Energy contribution | -3.52 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.05 |
| Structure conservation index | 0.12 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.993307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
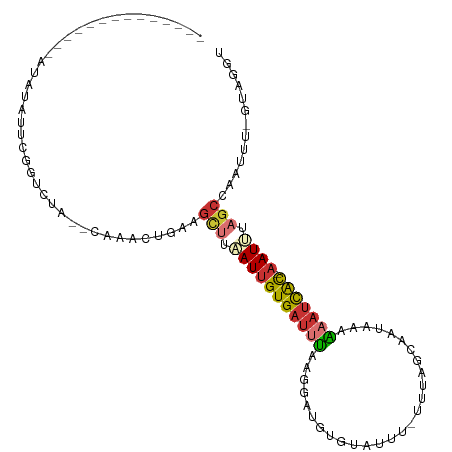
>dm3.chr3R 20518222 91 - 27905053 ACCUAC-AAAGUGGCUAAAUU-GUGAUUUUUAACUGCUAAA-AAAUACACAUCCAUAAAUCACAAUUAAGCUUCAGUUUG--UAGACCGAAUAAAU-------------- ..((((-(((.(((((.((((-(((((((.....((.....-.......)).....))))))))))).)))..)).))))--)))...........-------------- ( -22.30, z-score = -4.86, R) >droPer1.super_7 2576082 105 + 4445127 -GUUGUACAGGAAGCUUGAUU-GUGAUUGG--AUUAUCCCA-GAACAUAUAGGCUCCAAUCACAAUGAAACUUCAUUUUGUAUAAACCAGAGAAACGUGUGUAAGUUUUU -.(((((((.((((.((.(((-((((((((--(....((..-.........)).)))))))))))).)).))))....))))))).....((((((........)))))) ( -28.80, z-score = -3.04, R) >dp4.chr2 29079681 104 + 30794189 -GUUGUACAGGAAGCUUGAUU-GUGAUUGG--AUUAUCCCA-AAAUAUA-AGGCUCCAAUCACAAUGAAACUUGAUUUUGUAUAAACCAGAGAAACGUGUGUAAGUUUUU -.((((((((((((.((.(((-((((((((--(....((..-.......-.)).)))))))))))).)).))...)))))))))).....((((((........)))))) ( -26.70, z-score = -2.89, R) >droEre2.scaffold_4820 2930076 90 + 10470090 GCCUAC-AAAUUGGCUAAAUU-GUGAUGUAUUACUACU-UG-AAAUAUACAUCCUUAAAUCACAAUUAUGCUUAUGUUUG--UAGGCCGAAUAUAU-------------- ((((((-((((.(((..((((-((((((((((.(....-.)-.)))))..........)))))))))..)))...)))))--))))).........-------------- ( -25.90, z-score = -5.11, R) >droYak2.chr3R 2853572 91 + 28832112 GCCUACAAAAUUGGCUUCAUU-GUGAUUUUUCGUUGCU-UG-AAAUUCACAUCCUUAAAUCACAAUUAAGCUCAAGUUUG--UAUGCCGAAUAUAU-------------- ((.((((((.(((((((.(((-(((((((.....((..-((-.....)))).....)))))))))).)))).))).))))--)).)).........-------------- ( -24.60, z-score = -4.60, R) >droSec1.super_0 20852382 91 - 21120651 ACCUAC-AAAUUGGCUAAAUU-GUGAUUUUUUAGUGCAAAA-AAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUG--UAGACCGAAUAUAU-------------- ..((((-(((((((((.((((-(((((((....(((.....-.....)))......))))))))))).)))..)))))))--)))...........-------------- ( -26.00, z-score = -6.08, R) >droSim1.chr3R_random 984822 91 - 1307089 ACCUAC-AAAUUGGCUAAAUU-GUGAUUUUUUAGUGCUAAA-AAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUG--UAGACCGAAUAUAU-------------- ..((((-(((((((((.((((-(((((((....(((.....-.....)))......))))))))))).)))..)))))))--)))...........-------------- ( -26.00, z-score = -6.07, R) >apiMel3.GroupUn 298547683 94 + 399230636 AUUUAAAAAGUGUGCGAGAUCAACAAUUAUAUAAAAACAGGCAGAGAAACAAAAAUCUCGCACUAUCAGUGUUCAUUUU---UCGAGUGUGUCCAAG------------- ...........(((((((((..................................))))))))).....(..(((.....---..)))..).......------------- ( -12.88, z-score = 0.26, R) >consensus ACCUAC_AAAUUGGCUAAAUU_GUGAUUUUUUAUUACUAAA_AAAUACACAUCCUUAAAUCACAAUUAAGCUUCAGUUUG__UAGACCGAAUAUAU______________ ......................(((((((...........................)))))))............................................... ( -2.83 = -3.52 + 0.69)

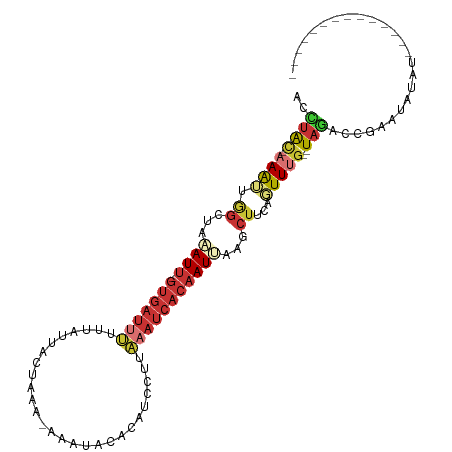
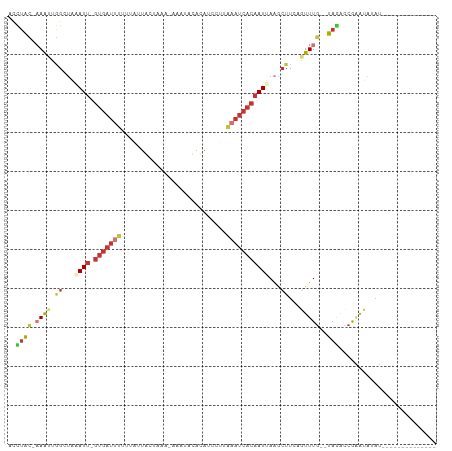
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:27 2011