| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,501,331 – 20,501,405 |
| Length | 74 |
| Max. P | 0.980906 |

| Location | 20,501,331 – 20,501,405 |
|---|---|
| Length | 74 |
| Sequences | 14 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Shannon entropy | 0.29668 |
| G+C content | 0.37589 |
| Mean single sequence MFE | -17.63 |
| Consensus MFE | -13.64 |
| Energy contribution | -14.96 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
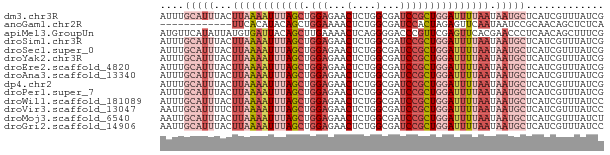
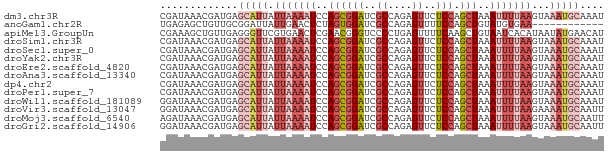
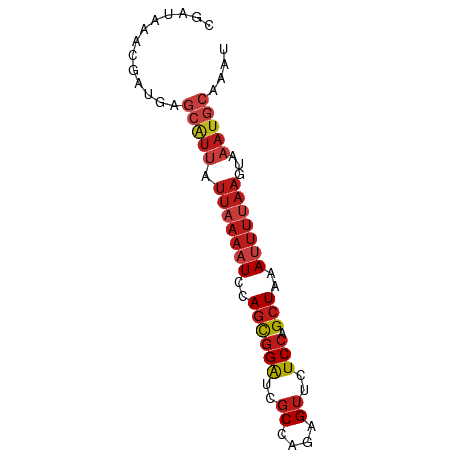
>dm3.chr3R 20501331 74 + 27905053 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >anoGam1.chr2R 44155550 62 + 62725911 ------------UUCACAUACAGCUGGAAAACUCUGGCGAUCCACUAGAGUUCAAUAAUCCGCAACAGCUCUCA ------------.........(((((...((((((((.......)))))))).............))))).... ( -11.99, z-score = -1.58, R) >apiMel3.GroupUn 14476131 74 + 399230636 AUGUUCAUAUUAUGUGAUUACAGCUUGAAAACUCAGGGGACCCGUUCGAGUUCACGAACCCUCAACAGCUUUCG .(((((((.....))))..)))((((((....)))((((....(((((......)))))))))...)))..... ( -12.80, z-score = 0.25, R) >droSim1.chr3R 20343957 74 + 27517382 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droSec1.super_0 20835804 74 + 21120651 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droYak2.chr3R 2836515 74 - 28832112 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droEre2.scaffold_4820 2913228 74 - 10470090 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droAna3.scaffold_13340 19023573 74 - 23697760 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >dp4.chr2 29060159 74 - 30794189 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droPer1.super_7 2557692 74 - 4445127 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.73, R) >droWil1.scaffold_181089 2772547 74 - 12369635 AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCC ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.71, R) >droVir3.scaffold_13047 799371 74 + 19223366 AAUUGCAUUUUCUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCC ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.53, R) >droMoj3.scaffold_6540 4800240 74 + 34148556 AAUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCU ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.85, R) >droGri2.scaffold_14906 8666472 74 - 14172833 AAUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCC ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. ( -18.50, z-score = -2.71, R) >consensus AUUUGCAUUUACUUAAAAUUUAGCUGGAGAACUCUGGCGAUCCGCUGGAUUUUAAUAAUGCUCAUCGUUUAUCG ....(((((...((((((((((((.(((...(....)...))))))))))))))).)))))............. (-13.64 = -14.96 + 1.32)
| Location | 20,501,331 – 20,501,405 |
|---|---|
| Length | 74 |
| Sequences | 14 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Shannon entropy | 0.29668 |
| G+C content | 0.37589 |
| Mean single sequence MFE | -15.38 |
| Consensus MFE | -10.71 |
| Energy contribution | -11.88 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
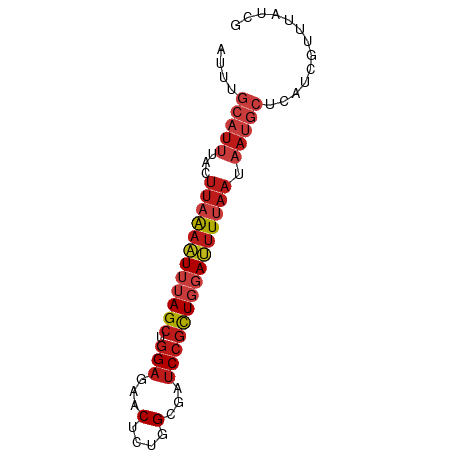
>dm3.chr3R 20501331 74 - 27905053 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >anoGam1.chr2R 44155550 62 - 62725911 UGAGAGCUGUUGCGGAUUAUUGAACUCUAGUGGAUCGCCAGAGUUUUCCAGCUGUAUGUGAA------------ .(((((((.(.((((.((((((.....)))))).)))).).)))))))..............------------ ( -14.60, z-score = -0.50, R) >apiMel3.GroupUn 14476131 74 - 399230636 CGAAAGCUGUUGAGGGUUCGUGAACUCGAACGGGUCCCCUGAGUUUUCAAGCUGUAAUCACAUAAUAUGAACAU .(((((((...(.(((..(((........)))..))).)..))))))).......................... ( -17.10, z-score = -0.56, R) >droSim1.chr3R 20343957 74 - 27517382 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droSec1.super_0 20835804 74 - 21120651 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droYak2.chr3R 2836515 74 + 28832112 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droEre2.scaffold_4820 2913228 74 + 10470090 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droAna3.scaffold_13340 19023573 74 + 23697760 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >dp4.chr2 29060159 74 + 30794189 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droPer1.super_7 2557692 74 + 4445127 CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.24, R) >droWil1.scaffold_181089 2772547 74 + 12369635 GGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -1.94, R) >droVir3.scaffold_13047 799371 74 - 19223366 GGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGAAAAUGCAAUU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -1.78, R) >droMoj3.scaffold_6540 4800240 74 - 34148556 AGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAUU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -2.32, R) >droGri2.scaffold_14906 8666472 74 + 14172833 GGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAUU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... ( -15.30, z-score = -1.83, R) >consensus CGAUAAACGAUGAGCAUUAUUAAAAUCCAGCGGAUCGCCAGAGUUCUCCAGCUAAAUUUUAAGUAAAUGCAAAU .............(((((.(((((((..((((((..((....))..))).)))..)))))))...))))).... (-10.71 = -11.88 + 1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:24 2011