| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,476,663 – 20,476,739 |
| Length | 76 |
| Max. P | 0.518481 |

| Location | 20,476,663 – 20,476,739 |
|---|---|
| Length | 76 |
| Sequences | 13 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 64.87 |
| Shannon entropy | 0.80975 |
| G+C content | 0.49096 |
| Mean single sequence MFE | -14.88 |
| Consensus MFE | -5.29 |
| Energy contribution | -5.18 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
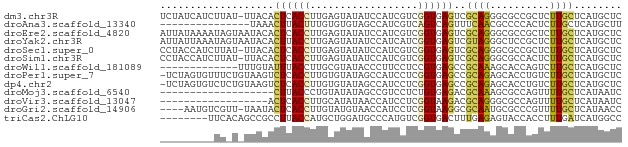
>dm3.chr3R 20476663 76 - 27905053 UCUAUCAUCUUAU-UUACACUCACCUUGAGUAUAUCCAUCGUCGGUGAGUCGCAGGGCGCCGCUCUUGCUCAUGCUC .............-....(((((((.(((.........)))..))))))).((((((((.......))))).))).. ( -18.20, z-score = -1.29, R) >droAna3.scaffold_13340 18999975 62 + 23697760 ---------------UAAACUUACUUUGUGUGUAGCCAUCGUCAGUCAGUUUCAACGCCCCACUCUUGCUCAUGCUU ---------------............(((((.(((....((......((....)).....))....)))))))).. ( -5.10, z-score = 1.07, R) >droEre2.scaffold_4820 2888101 77 + 10470090 AUUAUAAAAUAGUAAUACACUCACCUUGAGUAUAUCCAUCGUCGGUGAGUCGCAGGGCGCCGCUCUUGCUCAUGCUC ..........((((....(((((((.(((.........)))..))))))).(((((((...)).)))))...)))). ( -18.50, z-score = -1.11, R) >droYak2.chr3R 2811034 77 + 28832112 AUUAUUAAAUAGUAAUACACUUACCUUGAGUAUAUCCAUCAUCGGUGAGUCGUAGGGCUCCGCUCUUGCUCAUGCUC ..........((((.((((((((((.(((.........)))..))))))).)))((((.........)))).)))). ( -17.10, z-score = -1.42, R) >droSec1.super_0 20811054 76 - 21120651 CCUACCAUCUUAU-UUACACUCACCUUGAGUAUAUCCAUCGUCGGUGAGUCGCAGGGCGCCGCUCUUGCUCAUGCUC .............-....(((((((.(((.........)))..))))))).((((((((.......))))).))).. ( -18.20, z-score = -1.19, R) >droSim1.chr3R 20315517 76 - 27517382 CCUACCAUCUUAU-UUACACUCACCUUGAGUAUAUCCAUCGUCGGUGAGUCGCAGGGCGCCACUCUUGCUCAUGCUC .............-....(((((((.(((.........)))..))))))).((((((((.......))))).))).. ( -18.20, z-score = -1.66, R) >droWil1.scaffold_181089 2745072 64 + 12369635 -------------UUUGUAUUUACCUUGCGUAUACCCUUCCUCCGUGAGCCGCAAAGCACCAGUCUUGCUCAUGCUC -------------...((((....((..((.............))..))..((((..(....)..))))..)))).. ( -9.42, z-score = -0.58, R) >droPer1.super_7 2531681 76 + 4445127 -UCUAGUGUUUCUGUAAGUCUCACCUUGUGUAUAGCCAUCCUCGGUGAGCCGCAGAGCACCUGUCUUGCUCAUGCUC -....(((((.((((..(.((((((..(((......)))....))))))).))))))))).......((....)).. ( -18.70, z-score = -0.77, R) >dp4.chr2 29034138 76 + 30794189 -UCUAGUGUCUCUGUAAGUCUCACCUUGUGUAUAGCCAUCCUCGGUGAGCCGCAGAGCACCUGUCUUGCUCAUGCUC -....(((.((((((..(.((((((..(((......)))....))))))).))))))))).......((....)).. ( -21.10, z-score = -1.45, R) >droMoj3.scaffold_6540 4767515 58 - 34148556 -------------------CUUACCCUGUAUAUAGCCGUCCUCUGUGAGACGCAAAGCGCCAGUUUUGCUCAUAAUC -------------------..................((((.....).)))(((((((....)))))))........ ( -11.00, z-score = -1.19, R) >droVir3.scaffold_13047 772505 59 - 19223366 ------------------ACUCACCUUGCAUAUAACCAUCCUCGGUAAGACGCAGGGCGCCAGUUUUGCUCAUAAUC ------------------((((.((((((.....(((......))).....)))))).)..)))............. ( -10.90, z-score = -0.49, R) >droGri2.scaffold_14906 8634032 72 + 14172833 ----AAUGUCGUU-UAAUACUCACCUUGUAUGUAACCAUCCUCGGUAAGGCGCAAUGCGCCCGUUUUGCUCAUAACC ----.(((..((.-.(((....(((..(.(((....))).)..)))..(((((...))))).)))..)).))).... ( -12.40, z-score = -0.35, R) >triCas2.ChLG10 6610320 69 + 8806720 --------UUCACAGCCGCCUUACCAUGCUGGAUGCCCAUGUCGGUGACUUUGAGAGUACCACCUUUGAUCAUGGCC --------.((.((((...........))))))...(((((((((((((((...))))..))))...)).))))).. ( -14.60, z-score = 0.27, R) >consensus _________UU_U_UUACACUCACCUUGAGUAUAGCCAUCCUCGGUGAGUCGCAGGGCGCCACUCUUGCUCAUGCUC ...................((((((..................))))))..((((..........))))........ ( -5.29 = -5.18 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:21 2011