| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,467,181 – 20,467,269 |
| Length | 88 |
| Max. P | 0.971491 |

| Location | 20,467,181 – 20,467,269 |
|---|---|
| Length | 88 |
| Sequences | 14 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 85.44 |
| Shannon entropy | 0.33161 |
| G+C content | 0.45455 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.33 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
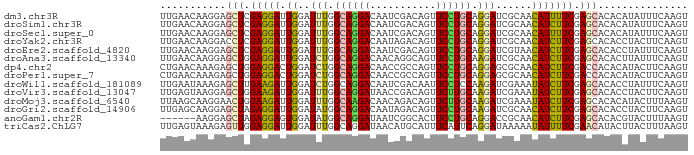
>dm3.chr3R 20467181 88 - 27905053 UUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUUUUCGAGCACACAUAUUUCAAGU (((((....(.((((((((((((((((..(((((((........).))))))..)))).))..)))))))))).)......))))).. ( -29.50, z-score = -4.41, R) >droSim1.chr3R 20306023 88 - 27517382 UUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUCUUCGAGCACACAUAUUUCAAGU (((((....(.((((((((((((((((..(((((((........).))))))..)))).))..)))))))))).)......))))).. ( -32.20, z-score = -4.91, R) >droSec1.super_0 20801546 88 - 21120651 UUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUUUUCGAGCACACAUAUUUCAAGU (((((....(.((((((((((((((((..(((((((........).))))))..)))).))..)))))))))).)......))))).. ( -29.50, z-score = -4.41, R) >droYak2.chr3R 2801533 88 + 28832112 UUGAACAAGGACCUCGAGGAUUGGAUUUGGCAGGACAAUAGACAGUUCCUGCAGGAUCGCAACAUCUUCGAGCACACCUACUUCAAGU (((((..(((..(((((((((((((((..(((((((........).))))))..)))).))..)))))))))....)))..))))).. ( -31.60, z-score = -4.33, R) >droEre2.scaffold_4820 2878573 88 + 10470090 UUGAACAAGGAGCUCGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGUAACAUCUUCGAGCACACCUAUUUCAAGU (((((..(((.((((((((((..((((..(((((((........).))))))..)))).....))))))))))...)))..))))).. ( -33.10, z-score = -4.59, R) >droAna3.scaffold_13340 18990521 88 + 23697760 UUGAACAAGGAGCUGGAGGAUUGGAUCUGGCAGGACAACAGGCAGUUCCUGCAAGAUCGCAACAUCUUCGAGCACACUUAUUUCAAGU (((((.((((.(((.(((((((((((((.(((((((........).)))))).))))).))..)))))).))).).)))..))))).. ( -32.40, z-score = -4.11, R) >dp4.chr2 29023569 88 + 30794189 CUGAACAAAGAGCUGGAGGACUGGAUCUGGCAGGACAACCGCCAGUUCCUGCAGGAGCGCAACAUCUUCGACCACACAUACUUCAAGU ...........(((.((((...(((.(((((.((....))))))).)))(((......)))...................)))).))) ( -21.60, z-score = 0.15, R) >droPer1.super_7 2520811 88 + 4445127 CUGAACAAAGAGCUGGAGGACUGGAUCUGGCAGGACAACCGCCAGUUCCUGCAGGAGCGCAACAUCUUCGACCACACAUACUUCAAGU ...........(((.((((...(((.(((((.((....))))))).)))(((......)))...................)))).))) ( -21.60, z-score = 0.15, R) >droWil1.scaffold_181089 2735047 88 + 12369635 UUGAAUAAAGAGCUUGAAGAUUGGAUCUGGCAGGACAAUCGACAAUUCCUCCAAGAUCGAAAUAUCUUCGAGCACACCUAUUUCAAGU (((((......(((((((((((.(((((((.((((...........)))))).))))).)...))))))))))........))))).. ( -26.24, z-score = -3.42, R) >droVir3.scaffold_13047 761051 88 - 19223366 UUGAGUAAGGAGCUGGAAGAUUGGAUUUGGCAGGAUAACCGACAGUUCUUGCAAGAUCGAAAUAUCUUCGAGCACACCUACUUCAAGU (((((((.((.(((.(((((((.(((((.((((((...........)))))).))))).)...)))))).)))...))))).)))).. ( -28.00, z-score = -3.15, R) >droMoj3.scaffold_6540 4756858 88 - 34148556 UUAAGCAAGGAACUGGAAGAUUGGAUUUGGCAAGACAACAGACAGUUCUUGCAAGAUCGAAAUAUCUUCGAGCACACAUACUUUAAGU ............((.(((((((.(((((.(((((((........).)))))).))))).)...)))))).))................ ( -19.20, z-score = -1.96, R) >droGri2.scaffold_14906 8622277 88 + 14172833 UUGAGCAAGGAGCUAGAGGAUUGGAUAUGGCAGGACAAUAGACAGUUCCUGCAAGAUCGCAACAUCUUCGAGCACACCUACUUCAAGU (((((..(((.(((.(((((((((((...(((((((........).))))))...))).))..)))))).)))...)))..))))).. ( -27.30, z-score = -2.90, R) >anoGam1.chr2R 19882198 82 + 62725911 ------AAGGAGCUAGAGGAGUGGAUAUGGCAGGAUAAUCGGCACUUCCUGCAGGACCGCAACAUCUUCGAGCACACGUACUUUAAGU ------.....(((.(((((((((.....((((((...........))))))....))))....))))).)))............... ( -22.90, z-score = -1.25, R) >triCas2.ChLG7 8790684 88 + 17478683 UUGAGUAAAGAGUUGGAGGAUUGGAUUUGGCAGGAUAACAUGCAUUUCAUUCAGGAUAAAAAUAUUUUCGAACAUACUUACUUUAAGU ..((((((..((((.(((((.((((....(((........)))..)))).))..((((....))))))).))).)..))))))..... ( -12.60, z-score = -0.07, R) >consensus UUGAACAAGGAGCUGGAGGAUUGGAUUUGGCAGGACAAUCGACAGUUCCUGCAGGAUCGCAACAUCUUCGAGCACACAUACUUCAAGU ...........(((.(((((.((..(((.((((((...........)))))).)))......))))))).)))............... (-14.90 = -14.33 + -0.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:19 2011