
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,561,025 – 8,561,142 |
| Length | 117 |
| Max. P | 0.883210 |

| Location | 8,561,025 – 8,561,119 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.98 |
| Shannon entropy | 0.56677 |
| G+C content | 0.50068 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.15 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
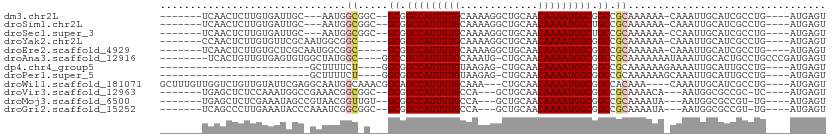
>dm3.chr2L 8561025 94 + 23011544 -------UCAACUCUUGUGAUUGC---AAUGGCGGC--GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAAAA-CAAAUUGCAUCGCCUG----AUGAGU -------...((((..(((((.((---(((.(((((--.((.((((((((((......))))...)))))))))))))......-...))))))))))...----..)))) ( -33.50, z-score = -1.86, R) >droSim1.chr2L 8342982 94 + 22036055 -------UCAACUCUUGUGAUUGC---AAUGGCGGC--GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCUGCCGCAAAAAA-CAAAUUGCAUCGCCUG----AUGAGU -------...((((..(((((.((---(((.(((((--..((((((((((((......))))...)))))))))))))......-...))))))))))...----..)))) ( -34.20, z-score = -2.40, R) >droSec1.super_3 4048226 94 + 7220098 -------UCAACUCUUGUGAUUGC---AAUGGCGGC--GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCUGCCGCAAAAAA-CCAAUUGCAUCGCCUG----AUGAGU -------...((((..(((((.((---(((((..((--((((((((((((((......))))...))))))))).)))......-)).))))))))))...----..)))) ( -34.20, z-score = -2.20, R) >droYak2.chr2L 11207605 94 + 22324452 -------CCAACUCUUGUGUUCGCAAUGGCGGC-----GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAAAA-CAAAUUGCAUCGCCUG----AUGAGU -------...((((((((....)))).(((((.-----((.(((((((((((......))))...))))))).)).((((....-....)))).)))))..----..)))) ( -31.10, z-score = -0.69, R) >droEre2.scaffold_4929 9156701 94 + 26641161 -------UCAACUCUUGUGCUCGCAAUGGCGGC-----GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAAAA-CAAAUUGCAUCGCCUG----AUGAGU -------...((((((((((.(((....)))))-----((((((...........)))))).))))...(((((..((((....-....)))).)))))..----..)))) ( -31.70, z-score = -0.94, R) >droAna3.scaffold_12916 10206469 98 + 16180835 --------UCACUGUUGUGAGUGUGGCUAUGGC----GGCCGCCAUUUUGCAAAUG-CUGCAACAAAAUGGCGGCCGCAAAAAAAUAAAUUGCACUGCCUGCCCGAUGAGU --------((((....)))).((.(((...(((----(((((((((((((((....-.))))...)))))))))))((((.........))))...))).)))))...... ( -40.70, z-score = -3.50, R) >dp4.chr4_group5 1314751 77 - 2436548 -------------------------GCUUUUCU----GGCGGCCAUUUUGUAAGAG-CUGCAACAAAAUGGCGGCCGCAAAAAAGAAAAUUGCAUUGCCUG----AUGAGU -------------------------((((.((.----(((.((((((((((.....-.....)))))))))).)))((((.........)))).......)----).)))) ( -25.70, z-score = -1.87, R) >droPer1.super_5 1284100 77 - 6813705 -------------------------GCUUUUCU----GGCGGCCAUUUUGUAAGAG-CUGCAACAAAAUGGCGGCCGCAAAAAAGCAAAUUGCAUUGCCUG----AUGAGU -------------------------((((((.(----(((.((((((((((.....-.....)))))))))).))))...))))))......((((....)----)))... ( -27.10, z-score = -1.79, R) >droWil1.scaffold_181071 974207 100 + 1211509 GCUUUGUUGGUCUGUUGUAUUCGAGGCAAUGGCAAACGGCAGCCAUUUUGCAAA---CUGCAACAAAAUGGCGGCCACAAA----CAAAUUGCAUCGCCUG----AUGAGU ..((((((.(((.(((((.......))))))))....(((.(((((((((....---......))))))))).)))...))----))))...((((....)----)))... ( -28.00, z-score = 0.13, R) >droVir3.scaffold_12963 14391469 91 - 20206255 -------UGAGCUCUCCAAAUGGCCGAAACGGCGGC--GCGGCCAUUUUGCCA---GCUGCAACAAAAUGGCGGCCGCAAAACA---AAUGGCGCCGC-UC----AUGAGU -------(((((..........((((...))))(((--((.(...((((((..---(((((.........))))).))))))..---..).)))))))-))----)..... ( -34.80, z-score = -0.84, R) >droMoj3.scaffold_6500 7409764 91 - 32352404 -------UGAGCUCUCGAAAUAGCCGUAACGGUUGU--GCGGCCAUUUUGCCA---GCUGCAACAAAAUGGCGGCCGCAAAAUA---AAUGGCGCCGU-UG----AUGAGU -------...((((((((....(((((....(((.(--((((((.....((((---..(......)..))))))))))).))).---.)))))....)-))----).)))) ( -33.50, z-score = -1.75, R) >droGri2.scaffold_15252 7637534 91 + 17193109 -------UCAGCCCUUGAAAUACCCAAAUCGGCGGC--GCGGCCAUUUUGCCA---GCUGCAACAAAAUGGCGGCCGCAAAAUA---AAUGGCGCCGU-UG----AUGAGU -------....................(((((((((--((.(..(((((((..---(((((.........))))).))))))).---..).)))))))-))----)).... ( -33.00, z-score = -1.22, R) >consensus _______UCAACUCUUGUGAUUGC_GCAAUGGC_G___GCGGCCAUUUUGCAAAAGGCUGCAACAAAAUGGCGGCCGCAAAAAA_CAAAUUGCAUCGCCUG____AUGAGU ......................................((.(((((((((.............))))))))).)).................................... (-14.15 = -14.15 + -0.00)


| Location | 8,561,025 – 8,561,119 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Shannon entropy | 0.56677 |
| G+C content | 0.50068 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
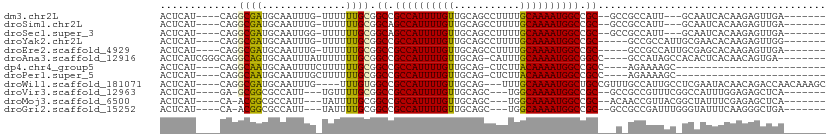
>dm3.chr2L 8561025 94 - 23011544 ACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC--GCCGCCAUU---GCAAUCACAAGAGUUGA------- ((((..----..((((.(((((....-....)))))((.((((((((((...........)))))))))).))--..)))).((---(......)))))))...------- ( -30.70, z-score = -1.19, R) >droSim1.chr2L 8342982 94 - 22036055 ACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC--GCCGCCAUU---GCAAUCACAAGAGUUGA------- ((((..----..((((.(((((....-....)))))((.((((((((((...........)))))))))).))--..)))).((---(......)))))))...------- ( -30.70, z-score = -1.21, R) >droSec1.super_3 4048226 94 - 7220098 ACUCAU----CAGGCGAUGCAAUUGG-UUUUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC--GCCGCCAUU---GCAAUCACAAGAGUUGA------- ((((..----..((((.(((((....-....)))))((.((((((((((...........)))))))))).))--..)))).((---(......)))))))...------- ( -30.70, z-score = -0.80, R) >droYak2.chr2L 11207605 94 - 22324452 ACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC-----GCCGCCAUUGCGAACACAAGAGUUGG------- ((((..----..((((.(((((....-....)))))((.((((((((((...........)))))))))).))-----..)))).(((......)))))))...------- ( -30.70, z-score = -0.82, R) >droEre2.scaffold_4929 9156701 94 - 26641161 ACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC-----GCCGCCAUUGCGAGCACAAGAGUUGA------- ((((..----...((..((((((...-......((((((((((((((((...........))))))))).)).-----))))).)))))).))....))))...------- ( -30.80, z-score = -0.62, R) >droAna3.scaffold_12916 10206469 98 - 16180835 ACUCAUCGGGCAGGCAGUGCAAUUUAUUUUUUUGCGGCCGCCAUUUUGUUGCAG-CAUUUGCAAAAUGGCGGCC----GCCAUAGCCACACUCACAACAGUGA-------- ........(((.(((...((((.........))))((((((((((((((.....-.....))))))))))))))----)))...))).((((......)))).-------- ( -41.20, z-score = -4.37, R) >dp4.chr4_group5 1314751 77 + 2436548 ACUCAU----CAGGCAAUGCAAUUUUCUUUUUUGCGGCCGCCAUUUUGUUGCAG-CUCUUACAAAAUGGCCGCC----AGAAAAGC------------------------- ......----...((...((((.........))))(((.((((((((((.....-.....)))))))))).)))----......))------------------------- ( -23.00, z-score = -1.93, R) >droPer1.super_5 1284100 77 + 6813705 ACUCAU----CAGGCAAUGCAAUUUGCUUUUUUGCGGCCGCCAUUUUGUUGCAG-CUCUUACAAAAUGGCCGCC----AGAAAAGC------------------------- .((..(----(..((((.((.....))....))))(((.((((((((((.....-.....)))))))))).)))----.))..)).------------------------- ( -25.00, z-score = -1.97, R) >droWil1.scaffold_181071 974207 100 - 1211509 ACUCAU----CAGGCGAUGCAAUUUG----UUUGUGGCCGCCAUUUUGUUGCAG---UUUGCAAAAUGGCUGCCGUUUGCCAUUGCCUCGAAUACAACAGACCAACAAAGC .....(----((((((((((((....----....((((.((((((((((.....---...)))))))))).)))).))).)))))))).)).................... ( -32.60, z-score = -2.01, R) >droVir3.scaffold_12963 14391469 91 + 20206255 ACUCAU----GA-GCGGCGCCAUU---UGUUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGC--GCCGCCGUUUCGGCCAUUUGGAGAGCUCA------- .....(----((-((..(.(((..---((......(((((((((((((((.....---.)))))))))).)).--)))((((...))))))..))).).)))))------- ( -37.00, z-score = -0.95, R) >droMoj3.scaffold_6500 7409764 91 + 32352404 ACUCAU----CA-ACGGCGCCAUU---UAUUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGC--ACAACCGUUACGGCUAUUUCGAGAGCUCA------- .(((..----.(-(((((((.(..---....).)))((.(((((((((((.....---.))))))))))).))--....))))).(((.....))).)))....------- ( -28.00, z-score = -1.07, R) >droGri2.scaffold_15252 7637534 91 - 17193109 ACUCAU----CA-ACGGCGCCAUU---UAUUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGC--GCCGCCGAUUUGGGUAUUUCAAGGGCUGA------- ......----..-.((((.((...---......(((((((((((((((((.....---.)))))))))).)).--)))))....((((.....)))))))))).------- ( -32.40, z-score = -0.51, R) >consensus ACUCAU____CAGGCGAUGCAAUUUG_UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGC___C_GCCAUUGC_GCAAUCACAAGAGUUGA_______ .............((((..............)))).((.((((((((((...........)))))))))).))...................................... (-15.57 = -15.13 + -0.44)
| Location | 8,561,046 – 8,561,142 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.47431 |
| G+C content | 0.52715 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.24 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
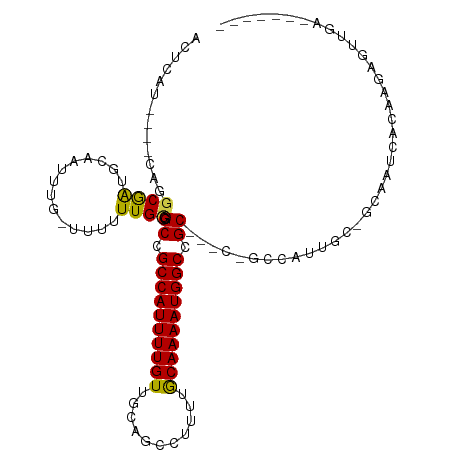
>dm3.chr2L 8561046 96 - 23011544 -----------------CGCACACCACC-----CACUCACUUAACACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGC-- -----------------...........-----..................----..(((..(((((....-....)))))((.((((((((((...........)))))))))).)))))..-- ( -26.00, z-score = -0.88, R) >droSim1.chr2L 8343003 79 - 22036055 ---------------------------------------CUUAACACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGC-- ---------------------------------------............----..(((..(((((....-....)))))((.((((((((((...........)))))))))).)))))..-- ( -26.00, z-score = -1.53, R) >droSec1.super_3 4048247 96 - 7220098 -----------------CACACACCACC-----CACUCACUUAACACUCAU----CAGGCGAUGCAAUUGG-UUUUUUGCGGCAGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGC-- -----------------...........-----..................----..(((..(((((....-....)))))((.((((((((((...........)))))))))).)))))..-- ( -26.00, z-score = -0.92, R) >droEre2.scaffold_4929 9156722 96 - 26641161 -----------------CACACACCACC-----CACUCACUUAACACUCAU----CAGGCGAUGCAAUUUG-UUUUUUGCGGCCGCCAUUUUGUUGCAGCCUUUUGCAAAAUGGCCGCGCCGC-- -----------------...........-----..................----..(((..(((((....-....)))))((.((((((((((...........)))))))))).)))))..-- ( -26.00, z-score = -1.49, R) >droAna3.scaffold_12916 10206492 115 - 16180835 CCGGGGCCAUCACAUCACCUCCUCACCC-----CACUCACUUAACACUCAUCGGGCAGGCAGUGCAAUUUAUUUUUUUGCGGCCGCCAUUUUGUUGCAGC-AUUUGCAAAAUGGCGGCCGC---- ..((((...................)))-----)...((((.....((.....)).....))))..............((((((((((((((((......-....))))))))))))))))---- ( -37.41, z-score = -2.11, R) >dp4.chr4_group5 1314761 90 + 2436548 -----------------CCCGGCCCGCC-----CACACACUUAACACUCAU----CAGGCAAUGCAAUUUUCUUUUUUGCGGCCGCCAUUUUGUUGCAGC-UCUUACAAAAUGGCCG-------- -----------------...((((.(((-----..................----..)))...((((.........))))))))((((((((((......-....))))))))))..-------- ( -26.05, z-score = -3.00, R) >droPer1.super_5 1284110 90 + 6813705 -----------------CCCGGCCCGCC-----CACACACUUAACACUCAU----CAGGCAAUGCAAUUUGCUUUUUUGCGGCCGCCAUUUUGUUGCAGC-UCUUACAAAAUGGCCG-------- -----------------...((((.(((-----..................----..)))...((((.........))))))))((((((((((......-....))))))))))..-------- ( -26.05, z-score = -2.14, R) >droWil1.scaffold_181071 974238 97 - 1211509 -----------------CAUCCGCCGCCAGCAACAUCCACUUAACACUCAU----CAGGCGAUGCAAUUUG----UUUGUGGCCGCCAUUUUGUUGCAGU---UUGCAAAAUGGCUGCCGUUUGC -----------------........((.(((....................----(((((((......)))----)))).(((.((((((((((......---..)))))))))).)))))).)) ( -28.30, z-score = -0.68, R) >droVir3.scaffold_12963 14391493 88 + 20206255 -----------------CAGUUGCCGCC-----CACU--GUUAACACUCAU----GA-GCGGCGCCAUUUG---UUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGCGCCGC-- -----------------..(.((((((.-----((.(--(........)))----).-)))))).).....---....(((((((((((((((((.....---.)))))))))).)).)))))-- ( -35.40, z-score = -1.39, R) >droMoj3.scaffold_6500 7409788 88 + 32352404 -----------------CACACACCGCC-----CACU--GUUAACACUCAU----CA-ACGGCGCCAUUUA---UUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGCACAAC-- -----------------.......((((-----...(--(...........----))-..)))).......---...(((((((...((((((((.....---.)))))))))))))))....-- ( -22.40, z-score = -0.38, R) >droGri2.scaffold_15252 7637558 85 - 17193109 --------------------AUGCCACA-----CAAU--GUUAACACUCAU----CA-ACGGCGCCAUUUA---UUUUGCGGCCGCCAUUUUGUUGCAGC---UGGCAAAAUGGCCGCGCCGC-- --------------------..(((...-----..((--(........)))----..-..)))........---....(((((((((((((((((.....---.)))))))))).)).)))))-- ( -27.50, z-score = -0.86, R) >consensus _________________CACACACCGCC_____CACUCACUUAACACUCAU____CAGGCGAUGCAAUUUG_UUUUUUGCGGCCGCCAUUUUGUUGCAGC_UUUUGCAAAAUGGCCGCGCCGC__ ..........................................................((((..............)))).((.((((((((((...........)))))))))).))....... (-15.52 = -15.24 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:20 2011