
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,455,805 – 20,455,899 |
| Length | 94 |
| Max. P | 0.572729 |

| Location | 20,455,805 – 20,455,899 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.51 |
| Shannon entropy | 0.47542 |
| G+C content | 0.50780 |
| Mean single sequence MFE | -22.62 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20455805 94 + 27905053 UCUCGGCAACCGUGUUUCCAUUUCCAGUUGACUAAUCG-CUCUGCCGACGUCGACGCAGGCCGC------------ACAACUGUUGCUGUUACUCUCCAACACCUUU ...(((((((.((((..((.......((((((...(((-......))).))))))...))..))------------))....))))))).................. ( -20.50, z-score = -0.54, R) >droSim1.chr3R 20294525 94 + 27517382 UCUCGGCAACCGUGUUUCCAUUUCCAGUUGACUAAUCG-CUCUGUCGACGUCGACGCAGGCCGU------------ACAACUGUUGCUGUUACUCUCCAACACCUUU ...(((((((.((.............((((((......-....))))))(((......)))...------------...)).))))))).................. ( -19.30, z-score = -0.23, R) >droSec1.super_0 20790206 94 + 21120651 UCUCGGCAACCGUGUUUCCAUUUCCAGUUGACUAAUCG-CUCUGCCGACGUCGACGCAGGCCGC------------ACAACUGUUGCUGUUACUCUCCAACACCUUU ...(((((((.((((..((.......((((((...(((-......))).))))))...))..))------------))....))))))).................. ( -20.50, z-score = -0.54, R) >droYak2.chr3R 2790080 88 - 28832112 UCUCGGCAACCGUGUUU------CCAGUUGACUAAUCG-CUCUGCCGACGUCGACGCAGCCCGC------------ACAACUGUUGCUGUUACUUUCCAACACCUUU ...(((((((.((....------...((((((...(((-......))).))))))((.....))------------...)).))))))).................. ( -19.90, z-score = -0.88, R) >droEre2.scaffold_4820 2867201 88 - 10470090 UCUCGGCAACCGUGUUU------CCAGUUGACUAAUCG-CUCUGCCGACGUCGACGCAGGCCAC------------ACAACUGUUGCUGUUACUUUCCAACACCUUU ...(((((((.((((..------((.((((((...(((-......))).))))))...))..))------------))....))))))).................. ( -22.00, z-score = -1.39, R) >dp4.chr2 29012138 89 - 30794189 UCUCAGCAACCGUUUCA---UUUCCAGUUGACUAAUCGGCUCUGCCGGCGUCGACGCUGGC---------------ACAACUGUUGCUGUUACUUUUCAAUUUCGUU ...(((((((.(((...---.....((((((....)))))).((((((((....)))))))---------------).))).))))))).................. ( -29.30, z-score = -3.91, R) >droPer1.super_7 2509318 89 - 4445127 UCUCAGCAACCGUUUCA---UUUCCAGUUGACUAAUCGGCUCUGCCGGCGUCGACGCUGGC---------------ACAACUGUUGCUGUUACUUUUCAAUUUCGUU ...(((((((.(((...---.....((((((....)))))).((((((((....)))))))---------------).))).))))))).................. ( -29.30, z-score = -3.91, R) >droWil1.scaffold_181089 2723147 79 - 12369635 UCUCGGCAACCAU--------UUCCAGUUGACUAAUCG-CUCUGCCGACUCCGACUGCGGCAGCG------UUAUGAGAACCUUUUGUGCUGUU------------- ((((((((((...--------.....)))).))....(-(.((((((.(.......))))))).)------)...))))...............------------- ( -18.50, z-score = -0.15, R) >droGri2.scaffold_14906 8605818 97 - 14172833 UCUCAGCAACGAU---------UCCAGUUGACUAAUCA-UGCUGCUUGCGUCGACGCUGGCGUCACUGUCAUUGUCACAACUGUUGCUGUUACUUUCCAUUUAUGUU ...((((((((..---------..)(((((.......(-(((.....)))).((((.(((((....))))).)))).)))))))))))).................. ( -24.30, z-score = -1.23, R) >consensus UCUCGGCAACCGUGUUU___UUUCCAGUUGACUAAUCG_CUCUGCCGACGUCGACGCAGGCCGC____________ACAACUGUUGCUGUUACUUUCCAACACCUUU ...(((((((.((.............((((((...(((.......))).))))))(....)..................)).))))))).................. (-11.60 = -11.96 + 0.36)
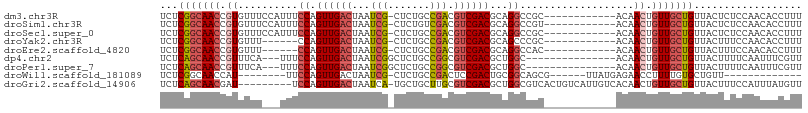
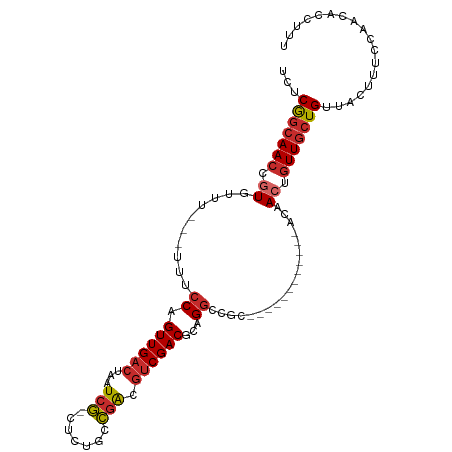

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:17 2011