
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,443,270 – 20,443,429 |
| Length | 159 |
| Max. P | 0.999172 |

| Location | 20,443,270 – 20,443,429 |
|---|---|
| Length | 159 |
| Sequences | 10 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.58101 |
| G+C content | 0.49127 |
| Mean single sequence MFE | -48.26 |
| Consensus MFE | -25.69 |
| Energy contribution | -25.15 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
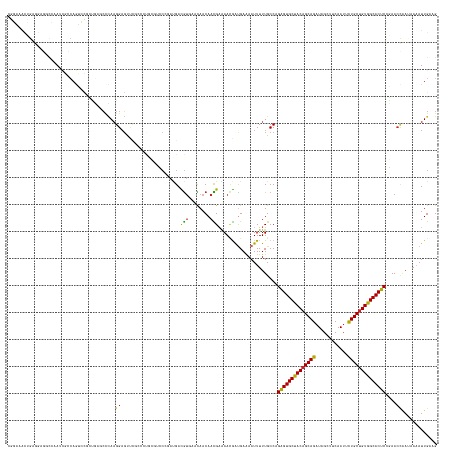
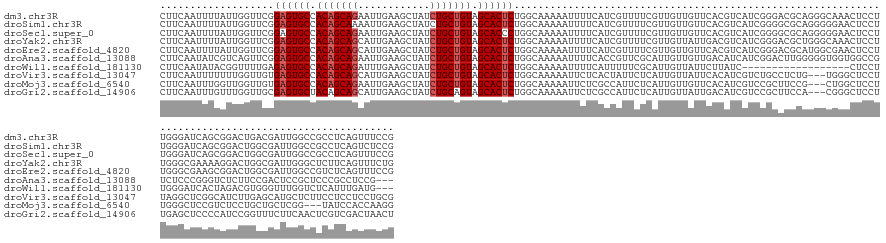
>dm3.chr3R 20443270 159 + 27905053 CGGAAACUGAGGCGGCCAAUCGUCAGUCCGCUGAUCCCAAGGAGUUUGCCCUGCGUCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG .((.......(((((....(((((..(((..((....)).)))((((.....(((((.....)))))))))............)))))......))))).(((((((((((((((..........)))))))))))))))....))............. ( -49.00, z-score = -2.59, R) >droSim1.chr3R 20281022 159 + 27517382 CGGAGACUGAGGCGGCCAAUCGCCAGUCCGCUGAUCCCAAGGAGUUCCCCCUGCGCCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUUUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG (((.((((..(((((....)))))))))(((((....))(((.......))))))..)))....(((........)))....((((.......((((...(((((((((((((((..........)))))))))))))))))))........))))... ( -43.86, z-score = -1.56, R) >droSec1.super_0 20776826 159 + 21120651 CGGAAACUGAGGCGGCCAAUCGCCAGUCCGCUGAUCCCAAGGAGUUCCCCCUGCGCCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGGGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG ((((......(((((....)))))..))))..........(((((...(((((.((........(((........))).....(((....)))...))))))).(((((((((((..........)))))))))))))))).................. ( -48.50, z-score = -2.23, R) >droYak2.chr3R 2776527 159 - 28832112 CAGAAACUGAAGAGCCCAAUCGCCAGUCCUUUUCGCCCAAGGAGUUUGCCCAGCGUCCCGAUGACGUCAAUAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG (((((((((..((......))..)))((.((((((.....((.(....))).(((((.....))))).........)))))).))......))))))...((((((((((((((.((.......))))))))))))))))................... ( -41.10, z-score = -2.15, R) >droEre2.scaffold_4820 2854631 159 - 10470090 CGGAAACUGAGACGGCCAAUCGCCAGUCCGCUUCGCCCAAGGAGUUCGCCAUGCGUCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG .((.....(.((((((.....))).))))..((((..((((((((((((....(((....)))..))))))..((.((....)).))....))))))...((((((((((((((.((.......))))))))))))))))))))))............. ( -46.20, z-score = -1.93, R) >droAna3.scaffold_13088 484505 156 + 569066 ---CGGAGGCGGGAGCGGAGUCGGAAGAGACCCGGGAGACGGCCACCACCCCCAAGUCCGAUGAUGUCAACAACAAUGCGAACGGUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACUGACGAUAUUGAAG ---.((.((.((..((((.(((......))))).(....).))..)).)).)).....(((((.(((((..............((..((....))..)).(((((((((((((((..........))))))))))))))).....))))).)))))... ( -61.40, z-score = -4.06, R) >droWil1.scaffold_181130 3411135 138 - 16660200 ---CAUCAAAUGAGACCAAACCCACGUCUAGUGAUCCCAAGGAG------------------GAUAAGAAUAACAAUGCGAAAAAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAAUCUGCUGUGGCACUCUCAAAACCGUAUAUUGAAG ---..((((......((.....(((.....))).(((...))))------------------)............(((((((((((....))))))...(((((((((((((((((........)))))))))))))))))......))))).)))).. ( -43.70, z-score = -5.60, R) >droVir3.scaffold_13047 15004545 156 - 19223366 CGCAGGAGGAGGAAGAGCAUGCUCAAGAUGCCGAGCCUAAGGAGCCCA---CAGAGGCAGACGAUGUGAAUAACAAUGAGAAUAGUGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUGCUGCUGUGGCACUCACAACCAAAAAAAUUGAAG ....((.(((((((...(((.((((.........((((..(.......---)..))))......(((.....))).))))....)))....))))).)).((((((((((((((.((.......))))))))))))))))....))............. ( -45.40, z-score = -1.99, R) >droMoj3.scaffold_6540 22132567 153 + 34148556 CCUUGGUGGAUA---CCGAGCAGCAGGAGACGGAGCCCAAGGAGCCAG---CGGAAGCGGACGAUGUGAACAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCACAACCAACCAAAUUGAAG ..(((((((...---(((..(.((.(....)((..((...))..)).)---).)...)))....(((...............(((((((....)))))))(((((((((((((((..........)))))))))))))))))).)).)))))....... ( -57.30, z-score = -4.57, R) >droGri2.scaffold_14906 4452109 156 - 14172833 AGUUAGUCGACGAGUUGAAGAAACCGGAUGGGGAGCUCAAGGAGCCCG---UGGAAGCGGACGAUGUCAAUAACAAUGAGAAUGGCGAGAAUUUUUGCCAGAGUGCUACUGCAGAUAGCUUCAAUGCUGCUGUAGCACUCGCAACCAAACAAAUUGAAG ......((((((.(((.....)))))..((.((.((((...)))))).---(((..((.(((...)))..............(((((((....)))))))(((((((((.((((.((.......)))))).)))))))))))..)))..))..)))).. ( -46.10, z-score = -1.34, R) >consensus CGGAAACUGAGGAGGCCAAUCGCCAGUCCGCUGAGCCCAAGGAGUUCGCCCUGCGUCCCGAUGACGUGAACAACAACGAAAACGAUGAAAAUUUUUGCCAGAGUGCUACAGCAGAUAGCUUCAAUUCUGCUGUGGCACUCCGAACCAAUAAAAUUGAAG ........................................((......................(((........)))......................((((((((((((((............))))))))))))))....))............. (-25.69 = -25.15 + -0.54)


| Location | 20,443,270 – 20,443,429 |
|---|---|
| Length | 159 |
| Sequences | 10 |
| Columns | 159 |
| Reading direction | reverse |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.58101 |
| G+C content | 0.49127 |
| Mean single sequence MFE | -47.10 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
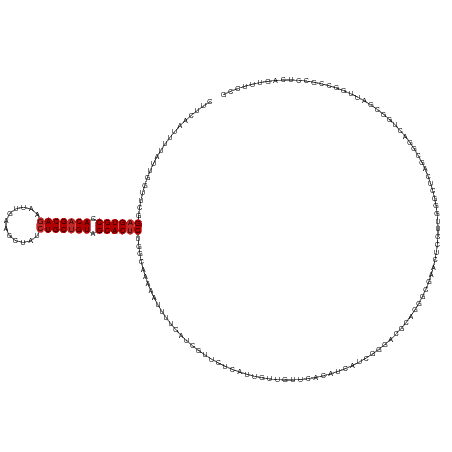
>dm3.chr3R 20443270 159 - 27905053 CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCACGUCAUCGGGACGCAGGGCAAACUCCUUGGGAUCAGCGGACUGACGAUUGGCCGCCUCAGUUUCCG .............((((((((((((.((((((((..........)))))))).))))))))............(((((((((((((..((((.((((.....))))(((((......))))))))))))))))..)))))).))))............. ( -51.70, z-score = -1.86, R) >droSim1.chr3R 20281022 159 - 27517382 CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAAAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCACGUCAUCGGGGCGCAGGGGGAACUCCUUGGGAUCAGCGGACUGGCGAUUGGCCGCCUCAGUCUCCG ............(((((((((((((.((((((..............)))))).)))))))................(((..(.((((.((((.((....)))))))))).)..)))......))))))(.(((((((((......))))..))))).). ( -50.34, z-score = -0.91, R) >droSec1.super_0 20776826 159 - 21120651 CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACCCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCACGUCAUCGGGGCGCAGGGGGAACUCCUUGGGAUCAGCGGACUGGCGAUUGGCCGCCUCAGUUUCCG .............((..(((.((((.((((((((..........)))))))).)))))).(((................(((((((..((((.((((.....))))((((((....)))))))))))))))))..(((.....))))))...)...)). ( -50.50, z-score = -0.48, R) >droYak2.chr3R 2776527 159 + 28832112 CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUAUUGACGUCAUCGGGACGCUGGGCAAACUCCUUGGGCGAAAAGGACUGGCGAUUGGGCUCUUCAGUUUCUG ..........(((((..((((((((.(((((((.((.......))))))))).))))))))((...((((.(((((.((((((((........((((.....))))(.(((......))).))))))))).)).))).))))..))...)))))..... ( -46.50, z-score = -0.81, R) >droEre2.scaffold_4820 2854631 159 + 10470090 CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUUUCGUUGUUGUUCACGUCAUCGGGACGCAUGGCGAACUCCUUGGGCGAAGCGGACUGGCGAUUGGCCGUCUCAGUUUCCG ..............((.((((((((.(((((((.((.......))))))))).)))))))))).......((((..(..(((((..((((((.((....))))))).)..)))))..)..)))).(((((((((.(((.....)))))))..))))).. ( -52.10, z-score = -1.53, R) >droAna3.scaffold_13088 484505 156 - 569066 CUUCAAUAUCGUCAGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCACCGUUCGCAUUGUUGUUGACAUCAUCGGACUUGGGGGUGGUGGCCGUCUCCCGGGUCUCUUCCGACUCCGCUCCCGCCUCCG--- ..........(((.((.((((((((.((((((((..........)))))))).)))))))))).............(((.(((....))).))).......)))..((((((((..((.(.....((((.....))))...).))..)))))))).--- ( -57.80, z-score = -3.18, R) >droWil1.scaffold_181130 3411135 138 + 16660200 CUUCAAUAUACGGUUUUGAGAGUGCCACAGCAGAUUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUUUUUCGCAUUGUUAUUCUUAUC------------------CUCCUUGGGAUCACUAGACGUGGGUUUGGUCUCAUUUGAUG--- ..((((.....((....((((((((.(((((((((........))))))))).))))))..((((((((....)))))).))........))....)------------------)....((((((((...(((....))))))))))).))))..--- ( -40.10, z-score = -2.69, R) >droVir3.scaffold_13047 15004545 156 + 19223366 CUUCAAUUUUUUUGGUUGUGAGUGCCACAGCAGCAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCACUAUUCUCAUUGUUAUUCACAUCGUCUGCCUCUG---UGGGCUCCUUAGGCUCGGCAUCUUGAGCAUGCUCUUCCUCCUCCUGCG ...(((((.....))))).((((((.(((((((.((.......))))))))).)))))).((((.................................(((..(....)---..))).......((((((....)))))).))))............... ( -36.30, z-score = 0.35, R) >droMoj3.scaffold_6540 22132567 153 - 34148556 CUUCAAUUUGGUUGGUUGUGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUGUUCACAUCGUCCGCUUCCG---CUGGCUCCUUGGGCUCCGUCUCCUGCUGCUCGG---UAUCCACCAAGG ......((((((.((((((((((((.((((((((..........)))))))).))))))..))))........(((.....................(((.((....)---).))).....((((..((.....))..))))))---)..)))))))). ( -43.90, z-score = -0.95, R) >droGri2.scaffold_14906 4452109 156 + 14172833 CUUCAAUUUGUUUGGUUGCGAGUGCUACAGCAGCAUUGAAGCUAUCUGCAGUAGCACUCUGGCAAAAAUUCUCGCCAUUCUCAUUGUUAUUGACAUCGUCCGCUUCCA---CGGGCUCCUUGAGCUCCCCAUCCGGUUUCUUCAACUCGUCGACUAACU ...(((((.....))))).(((((((((.((((.((.......)))))).)))))))))((((..........))))........(((((((((...(((((......---)))))...(((((....((....))....)))))...))))).)))). ( -41.80, z-score = -1.86, R) >consensus CUUCAAUUUUAUUGGUUCGGAGUGCCACAGCAGAAUUGAAGCUAUCUGCUGUAGCACUCUGGCAAAAAUUUUCAUCGUUCUCAUUGUUGUUCACAUCAUCGGGACGCAGGGCGAACUCCUUGGGCUCAGCGGACUGGCGAUUGGCCGCCUCAGUUUCCG ...................((((((.(((((((............))))))).)))))).................................................................................................... (-19.52 = -19.82 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:14 2011