
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,425,172 – 20,425,264 |
| Length | 92 |
| Max. P | 0.729893 |

| Location | 20,425,172 – 20,425,262 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Shannon entropy | 0.39777 |
| G+C content | 0.46222 |
| Mean single sequence MFE | -17.85 |
| Consensus MFE | -9.78 |
| Energy contribution | -11.58 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
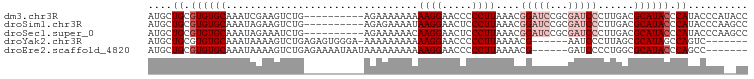
>dm3.chr3R 20425172 90 - 27905053 AUGCUGCGUGUGCAAAUCGAAGUCUG----------AGAAAAAAAAAGGAACCCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAUACC ....((.((((((........(((.(----------((.......((((.....))))....(((((...))))))))))))))))).)).......... ( -16.90, z-score = -1.71, R) >droSim1.chr3R 20263384 90 - 27517382 AUGCUGCGUGUGCAAAUAGAAGUCUG----------AGAGAAAAUAAGGAACUCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAAGCC ..(((..(((((....((...(((.(----------((......(((((.....)))))...(((((...)))))))))))...))..)))))...))). ( -19.30, z-score = -1.71, R) >droSec1.super_0 20759360 90 - 21120651 AUGCUGCGUGUGCAAAUAGAAAUCUG----------AGAAAAAACAAGGAACUCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAAGCC ....((.((((((.............----------.........((((.....))))....(((((...)))))......)))))).)).......... ( -16.60, z-score = -1.41, R) >droYak2.chr3R 2757854 86 + 28832112 AUGCUGCGUGUGCAAAUAAAAGUCUGAGAGUGGGA-AAAAAAAAAAAGGAACCCCCUUAAAACG------AAUCCCUUAGCGCAUAGCCAGUC------- ..(((((((((((...........(....).((((-.........((((.....))))......------..))))...)))))).).)))).------- ( -17.13, z-score = -1.18, R) >droEre2.scaffold_4820 2836943 87 + 10470090 AUGCUGCGUGUGCAAAUAAAAGUCUGAGAAAAUAAUAAAAAAAAAAAGGAACCCCCUUAAAACG------GAUCCCCUGGCGCAUACCCAGCC------- ..((((.((((((................................((((.....))))....((------(.....)))..)))))).)))).------- ( -19.30, z-score = -2.89, R) >consensus AUGCUGCGUGUGCAAAUAGAAGUCUG__________AGAAAAAAAAAGGAACCCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCA__CC ....((.((((((................................((((.....))))....(((((...)))))......)))))).)).......... ( -9.78 = -11.58 + 1.80)


| Location | 20,425,174 – 20,425,264 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Shannon entropy | 0.37435 |
| G+C content | 0.45325 |
| Mean single sequence MFE | -16.75 |
| Consensus MFE | -11.42 |
| Energy contribution | -12.36 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.729893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
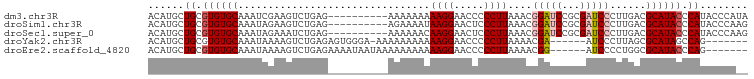
>dm3.chr3R 20425174 90 - 27905053 ACAUGCUGCGUGUGCAAAUCGAAGUCUGAG----------AAAAAAAAAGGAACCCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAUA ..(((((((....))).......(((.(((----------.......((((.....))))....(((((...)))))))))))))))............. ( -17.00, z-score = -1.52, R) >droSim1.chr3R 20263386 90 - 27517382 ACAUGCUGCGUGUGCAAAUAGAAGUCUGAG----------AGAAAAUAAGGAACUCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAAG ......((.((((((...(((....))).(----------((....(((((.....)))))...(((((...))))))))...)))))).))........ ( -18.90, z-score = -1.81, R) >droSec1.super_0 20759362 90 - 21120651 ACAUGCUGCGUGUGCAAAUAGAAAUCUGAG----------AAAAAACAAGGAACUCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCAAG ......((.((((((...............----------.......((((.....))))....(((((...)))))......)))))).))........ ( -16.60, z-score = -1.74, R) >droYak2.chr3R 2757856 86 + 28832112 ACAUGCUGCGUGUGCAAAUAAAAGUCUGAGAGUGGGA-AAAAAAAAAAAGGAACCCCCUUAAAACGA------AUCCCUUAGCGCAUAGCCAG------- ....(((..((((((...........(....).((((-.........((((.....)))).......------.))))...)))))))))...------- ( -15.93, z-score = -0.73, R) >droEre2.scaffold_4820 2836945 87 + 10470090 ACAUGCUGCGUGUGCAAAUAAAAGUCUGAGAAAAUAAUAAAAAAAAAAAGGAACCCCCUUAAAACGG------AUCCCCUGGCGCAUACCCAG------- .....(((.((((((................................((((.....))))....(((------.....)))..)))))).)))------- ( -15.30, z-score = -1.59, R) >consensus ACAUGCUGCGUGUGCAAAUAGAAGUCUGAG__________AAAAAAAAAGGAACCCCCUUAAACGGAUCCGCGAUCCCUUGACGCAUACCCAUACCCA__ ......((.((((((................................((((.....))))....(((((...)))))......)))))).))........ (-11.42 = -12.36 + 0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:08 2011