
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,424,859 – 20,424,971 |
| Length | 112 |
| Max. P | 0.664121 |

| Location | 20,424,859 – 20,424,971 |
|---|---|
| Length | 112 |
| Sequences | 7 |
| Columns | 134 |
| Reading direction | reverse |
| Mean pairwise identity | 58.82 |
| Shannon entropy | 0.75122 |
| G+C content | 0.48750 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -7.64 |
| Energy contribution | -9.29 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.22 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
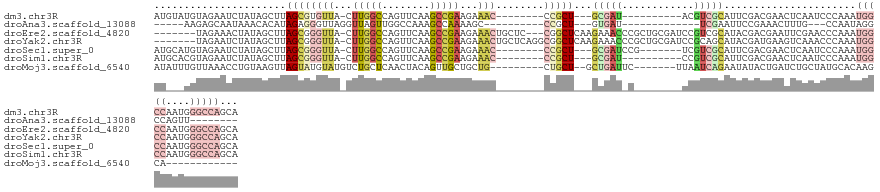
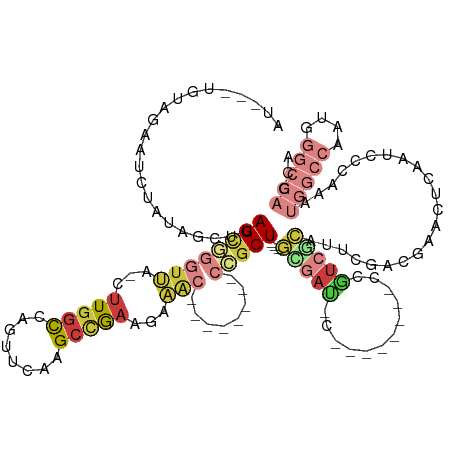
>dm3.chr3R 20424859 112 - 27905053 AUGUAUGUAGAAUCUAUAGCUUAGCGUGUUA-CUUGGCCAGUUCAAGCCGAAGAAAC--------CCGCU---GCGAU----------ACGUCGCAUUCGACGAACUCAAUCCCAAAUGGCCAAUGGGCCAGCA .....(((((...)))))((.(((((.(((.-((((((........))).))).)))--------.))))---)....----------.(((((....)))))..............(((((....))))))). ( -30.80, z-score = -0.91, R) >droAna3.scaffold_13088 466585 92 - 569066 -----AAGAGCAAUAAACACAUAGAGGGUUAGGUUAGUUGGCCAAAGCCAAAAGC----------CCGCU---GUGAU-------------UCGAAUUCCGAAACUUUG---CCAAUAGGCCAGUU-------- -----.((((.........(((((.(((((.......(((((....))))).)))----------)).))---))).(-------------(((.....)))).))))(---((....))).....-------- ( -23.90, z-score = -1.47, R) >droEre2.scaffold_4820 2836629 123 + 10470090 -------UAGAAACUAUAGCUUAGCGGGUUA-CUUGGCCAGUUCAAGCCGAAGAAACUGCUC---CGGCUCAAGAAACCCGCUGCGAUCCGUCGCAUACGACGAAUUCGAACCCAAAUGGCCAAUGGGCCAGCA -------...........((.(((((((((.-((((.........(((((.((......)).---))))))))).)))))))))(((..(((((....)))))...)))........(((((....))))))). ( -44.50, z-score = -3.28, R) >droYak2.chr3R 2757533 126 + 28832112 -------UAGAAUCUAUAGCUUAGCGGGUUA-CUUGGCCAGUUCAAGCCGAAGAAACUGCUCAGGCGGCUCAAGAAACCCGCUGCGAUCCGCAGCAUACGAUGAAGUCAAACCCAAAUGGCCAAUGGGCCAGCA -------....((((((.(((.((((((((.-(((((((.(((..(((.(......).)))..)))))).)))).))))))))(((...))))))))).)))...............(((((....)))))... ( -42.80, z-score = -1.84, R) >droSec1.super_0 20759038 115 - 21120651 AUGCAUGUAGAAUCUAUAGCUUAGCGGGUUA-CUUGGCCAGUUCAAGCCGAAGAAAC--------CCGCU---GCGAUCCG-------UCGUCGCAUUCGACGAACUCAAUCCCAAAUGGCCAAUGGGCCAGCA ..((.(((((...)))))))..((((((((.-((((((........))).))).)))--------)))))---((((....-------((((((....))))))..)).........(((((....))))))). ( -42.00, z-score = -3.09, R) >droSim1.chr3R 20263069 112 - 27517382 AUGCACGUAGAAUCUAUAGCUUAGCGGGUUA-CUUGGCCAGUUCAAGCCGAAGAAAC--------CCGCU---GCGAU----------CCGUCGCAUUCGACGAACUCAAUCCCAAAUGGCCAAUGGGCCAGCA .(((.(((.((((.........((((((((.-((((((........))).))).)))--------)))))---((((.----------...)))))))).)))..............(((((....)))))))) ( -41.10, z-score = -3.24, R) >droMoj3.scaffold_6540 14936502 104 + 34148556 AUAUUUGUUAAACCUGUAAGUUAGUAUGUAUGUCUGCUCAACUACAGUUGCUGCUG---------CUGCU--GCUGAUUC-------UUAAUCAGAAUAUACUGAUCUGCUAUGCACAAGCA------------ ...(((((.......(((..(((((((((...............((((.((....)---------).)))--)(((((..-------...))))).)))))))))..))).....)))))..------------ ( -23.00, z-score = -0.08, R) >consensus AU___UGUAGAAUCUAUAGCUUAGCGGGUUA_CUUGGCCAGUUCAAGCCGAAGAAAC________CCGCU___GCGAU_C________CCGUCGCAUUCGACGAACUCAAUCCCAAAUGGCCAAUGGGCCAGCA ..................(((((...(((((..(((((........)))))......................(((((............)))))......................)))))..)))))..... ( -7.64 = -9.29 + 1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:41:07 2011