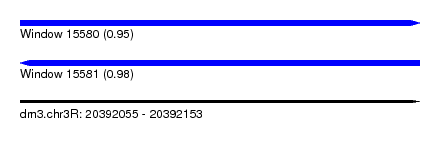
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,392,055 – 20,392,153 |
| Length | 98 |
| Max. P | 0.982303 |

| Location | 20,392,055 – 20,392,153 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 89.89 |
| Shannon entropy | 0.17857 |
| G+C content | 0.34758 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
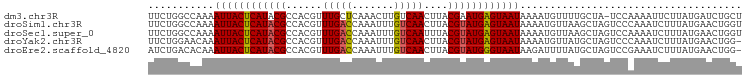
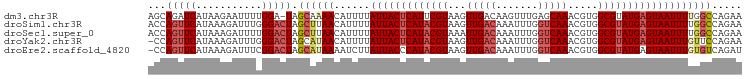
>dm3.chr3R 20392055 98 + 27905053 UUCUGGCCAAAAUUACUCAUACGCCACGUUUGCUCAAACUUGUCAACUUACGAAUGAGUAAUAAAAUGUUUUGCUA-UCCAAAAUUCUUAUGAUCUGCU ....(((....(((((((((.((....(((.((........)).)))...)).))))))))).....((((((...-..))))))...........))) ( -15.10, z-score = -1.42, R) >droSim1.chr3R 20230567 99 + 27517382 UUCUGGCCAAAAUUACUCAUACGCCACGUUUGACCAAAUUUGUCAACUUACGUAUGAGUAAUAAAAUGUUAAGCUAGUCCCAAAUCUUUAUGAACUGGU ..(((((((..((((((((((((......(((((.......)))))....))))))))))))....))....)))))..(((..((.....))..))). ( -23.30, z-score = -3.28, R) >droSec1.super_0 20727092 99 + 21120651 UUCUGGCCAAAAUUACUCAUACGCCACGUUUGACCAAAUUUGUCAAUUUACGUAUGAGUAAUAAAAUGUUAAGCUAGUCCAAAAUCUUUAUGAACUGGU ...((((....((((((((((((......(((((.......)))))....)))))))))))).....)))).((((((.((.........)).)))))) ( -23.50, z-score = -3.38, R) >droYak2.chr3R 2724546 98 - 28832112 UUCUGGAACAAAUUACUCAUACGCCACGUUUGACCAAAUUUGUCAACUUACGUAUGAGUAAUAAAAUGUUAUGCUAGUCCCAAAUCUUUAUGAACUGG- ..((((((((.((((((((((((......(((((.......)))))....))))))))))))....))))...))))..(((..((.....))..)))- ( -22.00, z-score = -3.05, R) >droEre2.scaffold_4820 2804615 98 - 10470090 AUCUGACACAAAUUACUCAUACGCCACGUUUGACCAAAUUUGUCAACUUACGUAUGGGUAAUAAGAUUUUAUGCUAGUCCGAAAUCUUUAUGAACUGG- ...........((((((((((((......(((((.......)))))....))))))))))))((((((((..........))))))))..........- ( -20.20, z-score = -1.91, R) >consensus UUCUGGCCAAAAUUACUCAUACGCCACGUUUGACCAAAUUUGUCAACUUACGUAUGAGUAAUAAAAUGUUAUGCUAGUCCAAAAUCUUUAUGAACUGGU ...........((((((((((((......(((((.......)))))....))))))))))))..................................... (-16.00 = -16.08 + 0.08)
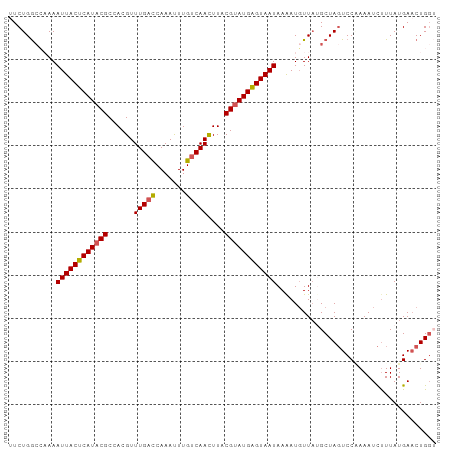
| Location | 20,392,055 – 20,392,153 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.89 |
| Shannon entropy | 0.17857 |
| G+C content | 0.34758 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.86 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
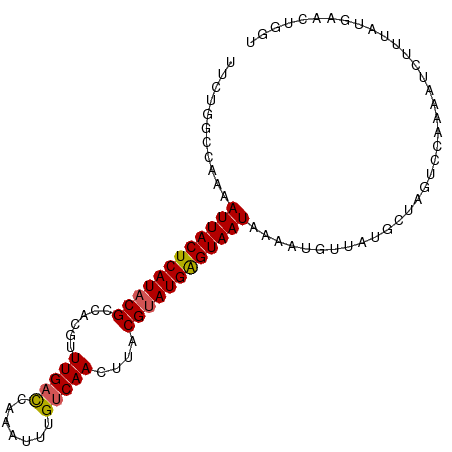
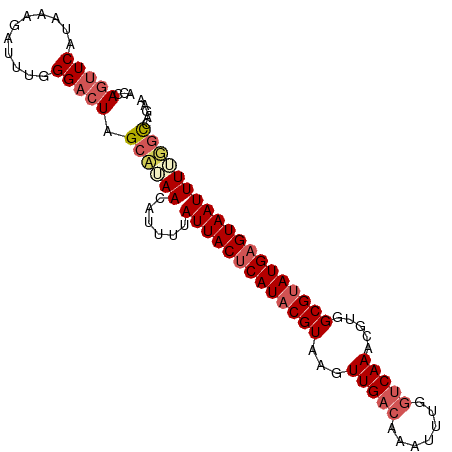
>dm3.chr3R 20392055 98 - 27905053 AGCAGAUCAUAAGAAUUUUGGA-UAGCAAAACAUUUUAUUACUCAUUCGUAAGUUGACAAGUUUGAGCAAACGUGGCGUAUGAGUAAUUUUGGCCAGAA ...............((((((.-...((((.......(((((((((.(((..(((..(........)..)))...))).))))))))))))).)))))) ( -18.51, z-score = -0.46, R) >droSim1.chr3R 20230567 99 - 27517382 ACCAGUUCAUAAAGAUUUGGGACUAGCUUAACAUUUUAUUACUCAUACGUAAGUUGACAAAUUUGGUCAAACGUGGCGUAUGAGUAAUUUUGGCCAGAA .((((.((.....)).))))..((.(((.........(((((((((((((..((((((.......))).)))...)))))))))))))...))).)).. ( -29.10, z-score = -3.79, R) >droSec1.super_0 20727092 99 - 21120651 ACCAGUUCAUAAAGAUUUUGGACUAGCUUAACAUUUUAUUACUCAUACGUAAAUUGACAAAUUUGGUCAAACGUGGCGUAUGAGUAAUUUUGGCCAGAA .((((.((.....))..)))).((.(((.........(((((((((((((...(((((.......))))).....)))))))))))))...))).)).. ( -26.70, z-score = -3.48, R) >droYak2.chr3R 2724546 98 + 28832112 -CCAGUUCAUAAAGAUUUGGGACUAGCAUAACAUUUUAUUACUCAUACGUAAGUUGACAAAUUUGGUCAAACGUGGCGUAUGAGUAAUUUGUUCCAGAA -..............((((((((..............(((((((((((((..((((((.......))).)))...)))))))))))))..)))))))). ( -29.89, z-score = -4.41, R) >droEre2.scaffold_4820 2804615 98 + 10470090 -CCAGUUCAUAAAGAUUUCGGACUAGCAUAAAAUCUUAUUACCCAUACGUAAGUUGACAAAUUUGGUCAAACGUGGCGUAUGAGUAAUUUGUGUCAGAU -..(((((...........))))).((((((......(((((.(((((((..((((((.......))).)))...))))))).)))))))))))..... ( -22.40, z-score = -1.88, R) >consensus ACCAGUUCAUAAAGAUUUGGGACUAGCAUAACAUUUUAUUACUCAUACGUAAGUUGACAAAUUUGGUCAAACGUGGCGUAUGAGUAAUUUUGGCCAGAA ...(((((...........))))).((((((......(((((((((((((...(((((.......))))).....)))))))))))))))))))..... (-20.50 = -21.86 + 1.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:57 2011