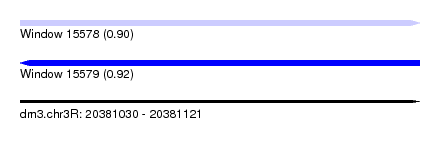
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,381,030 – 20,381,121 |
| Length | 91 |
| Max. P | 0.922907 |

| Location | 20,381,030 – 20,381,121 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Shannon entropy | 0.56718 |
| G+C content | 0.49965 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.27 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
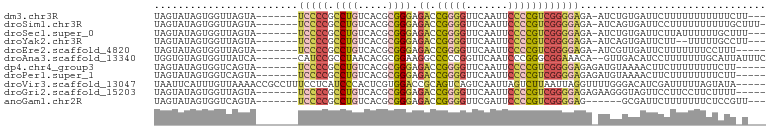
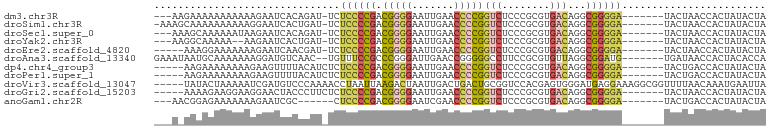
>dm3.chr3R 20381030 91 + 27905053 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUCUGUGAUUCUUUUUUUUUUUCUU--- .................-------(((((.((((.....)))).(((.((((.......))))))))))))((-(((...)))))..............--- ( -28.10, z-score = -1.02, R) >droSim1.chr3R 20219686 93 + 27517382 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUCAGUGAUUCCUUUUUUUUUUGCUUU- .....((.(((((....-------(((((.((((.....)))).(((.((((.......)))))))))))).)-)))).))....................- ( -30.40, z-score = -1.50, R) >droSec1.super_0 20716362 91 + 21120651 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUCUGUGAUUCUUAUUUUUUGCUUU--- ......((..(..((((-------(((((.((((.....)))).(((.((((.......))))))))))))((-(((...))))).))))..)..))..--- ( -32.10, z-score = -2.07, R) >droYak2.chr3R 2713260 89 - 28832112 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUCAGUGAUUCUU--UUUUUGCCUU--- .....((.(((((....-------(((((.((((.....)))).(((.((((.......)))))))))))).)-)))).))......--..........--- ( -30.40, z-score = -1.53, R) >droEre2.scaffold_4820 2794253 89 - 10470090 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUCGUUGAUUCUUUUUUUCCUUU----- .........((......-------(((((.((((.....)))).(((.((((.......))))))))))))((-(((...))))).......))...----- ( -28.70, z-score = -1.07, R) >droAna3.scaffold_13340 17948116 93 - 23697760 UGGUGUAGUGGUUAUCA-------CAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAACA--GUUGACAUCCUUUUUUUGCAUUAUUUC ((((((((.((...(((-------(.(((((.......))))).(.(((((((.......))))).))...).--).)))...)).....)))))))).... ( -26.10, z-score = -0.59, R) >dp4.chr4_group3 4659856 90 - 11692001 UAGUAUAGUGGUCAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUAAAACUUCUUUUUUUUCUU----- .((...(((...((...-------(((((.((((.....)))).(((.((((.......))))))))))))....))....))).))..........----- ( -27.80, z-score = -0.83, R) >droPer1.super_1 1798474 90 - 10282868 UAGUAUAGUGGUCAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUAAAACUUCUUUUUUUUCUU----- .((...(((...((...-------(((((.((((.....)))).(((.((((.......))))))))))))....))....))).))..........----- ( -27.80, z-score = -0.83, R) >droVir3.scaffold_13047 14934212 97 - 19223366 UAAUUCAUUUGUUAAAACCGCCUUUCCUCAUCCCACUCGUGGACCGCAGUCAGUCAAUUAGUCUUAAUUAGGUUUUGGGACAUCGAUUUUUAGUAUA----- .((((.((.(.((((((((((...(((.(.........).)))..))........(((((....))))).)))))))).).)).)))).........----- ( -10.60, z-score = 1.63, R) >droGri2.scaffold_15203 11499568 90 + 11997470 UAGUAUAGUGGUUAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAAGGGUAGUUCCUUCCUUCUUUU----- .................-------(((((.((((.....)))).(((.((((.......))))))))))))((((((((........))))))))..----- ( -34.60, z-score = -1.68, R) >anoGam1.chr2R 56631811 86 - 62725911 UAGUAUAGUGGUCAGUA-------UCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGGAG------GCGAUUCUUUUUUUCUCCGUU--- .........((((.((.-------(((((.((((.....)))).(((.((((.......)))))))))))).------))))))...............--- ( -28.50, z-score = 0.04, R) >consensus UAGUAUAGUGGUUAGUA_______UCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA_AUCUGUGAUUCUUUUUUUUUUUU_U___ ........................(((((.((((.....)))).((.(((((.......))))))))))))............................... (-18.91 = -19.27 + 0.36)
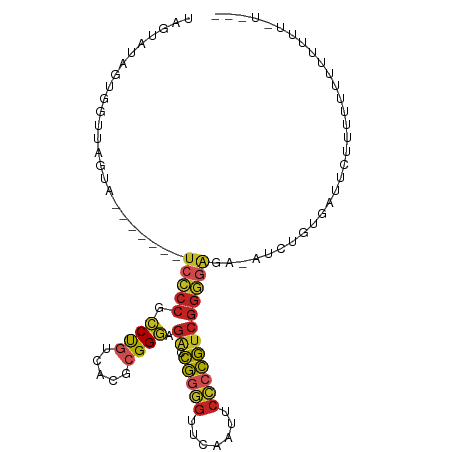
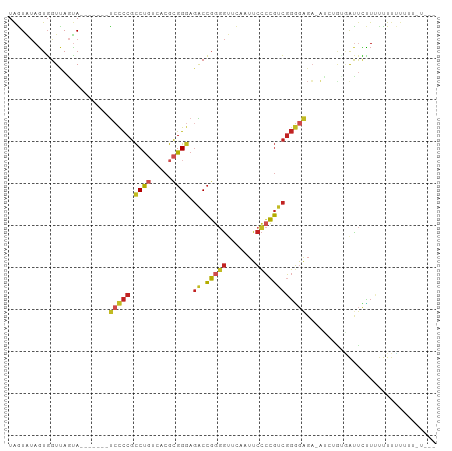
| Location | 20,381,030 – 20,381,121 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.27 |
| Shannon entropy | 0.56718 |
| G+C content | 0.49965 |
| Mean single sequence MFE | -25.59 |
| Consensus MFE | -17.32 |
| Energy contribution | -17.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.922907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
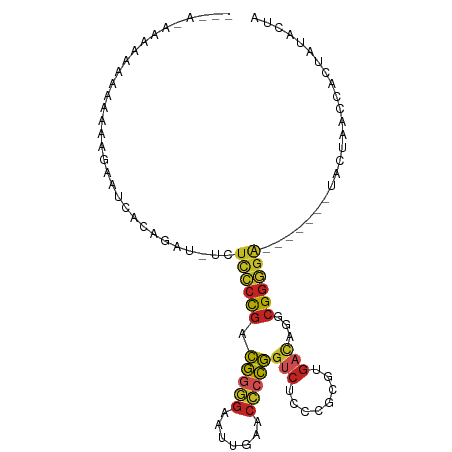
>dm3.chr3R 20381030 91 - 27905053 ---AAGAAAAAAAAAAAGAAUCACAGAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA ---..............(((((...)))-))((((((.(((((.......)))))(((........)))...))))))-------................. ( -26.10, z-score = -1.73, R) >droSim1.chr3R 20219686 93 - 27517382 -AAAGCAAAAAAAAAAGGAAUCACUGAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA -................(((((...)))-))((((((.(((((.......)))))(((........)))...))))))-------................. ( -25.90, z-score = -1.06, R) >droSec1.super_0 20716362 91 - 21120651 ---AAAGCAAAAAAUAAGAAUCACAGAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA ---..............(((((...)))-))((((((.(((((.......)))))(((........)))...))))))-------................. ( -26.10, z-score = -1.65, R) >droYak2.chr3R 2713260 89 + 28832112 ---AAGGCAAAAA--AAGAAUCACUGAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA ---..((......--..(((((...)))-))((((((.(((((.......)))))(((........)))...))))))-------......))......... ( -27.60, z-score = -1.59, R) >droEre2.scaffold_4820 2794253 89 + 10470090 -----AAAGGAAAAAAAGAAUCAACGAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA -----...((.......(((((...)))-))((((((.(((((.......)))))(((........)))...))))))-------......))......... ( -28.30, z-score = -1.80, R) >droAna3.scaffold_13340 17948116 93 + 23697760 GAAAUAAUGCAAAAAAAGGAUGUCAAC--UGUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUG-------UGAUAACCACUACACCA .................((.(((((..--....(((.(((((.......))))).((((..(....)..)))))))..-------))))).))......... ( -26.50, z-score = -0.33, R) >dp4.chr4_group3 4659856 90 + 11692001 -----AAGAAAAAAAAGAAGUUUUACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUGACCACUAUACUA -----.............(((......((..((((((.(((((.......)))))(((........)))...))))))-------....)).......))). ( -23.62, z-score = -0.26, R) >droPer1.super_1 1798474 90 + 10282868 -----AAGAAAAAAAAGAAGUUUUACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUGACCACUAUACUA -----.............(((......((..((((((.(((((.......)))))(((........)))...))))))-------....)).......))). ( -23.62, z-score = -0.26, R) >droVir3.scaffold_13047 14934212 97 + 19223366 -----UAUACUAAAAAUCGAUGUCCCAAAACCUAAUUAAGACUAAUUGACUGACUGCGGUCCACGAGUGGGAUGAGGAAAGGCGGUUUUAACAAAUGAAUUA -----.......(((((((...(((((.....((((((....)))))).((.(((.((.....))))).)).)).)))....)))))))............. ( -17.30, z-score = -0.15, R) >droGri2.scaffold_15203 11499568 90 - 11997470 -----AAAAGAAGGAAGGAACUACCCUUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUAACCACUAUACUA -----.......((((((......)))))).((((((.(((((.......)))))(((........)))...))))))-------................. ( -30.40, z-score = -1.34, R) >anoGam1.chr2R 56631811 86 + 62725911 ---AACGGAGAAAAAAAGAAUCGC------CUCCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACAGGCGGGGA-------UACUGACCACUAUACUA ---...((............(((.------.(((((...)))))..)))....((((.((((.(((.....)))))))-------.)))).))......... ( -26.00, z-score = -0.22, R) >consensus ___A_AAAAAAAAAAAAGAAUCACAGAU_UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGA_______UACUAACCACUAUACUA ...............................((((((.(((((.......)))))(((........)))...))))))........................ (-17.32 = -17.57 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:55 2011