
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,541,760 – 8,541,847 |
| Length | 87 |
| Max. P | 0.959413 |

| Location | 8,541,760 – 8,541,847 |
|---|---|
| Length | 87 |
| Sequences | 10 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 83.41 |
| Shannon entropy | 0.37397 |
| G+C content | 0.54810 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -30.53 |
| Energy contribution | -29.56 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
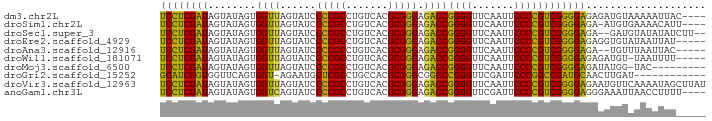
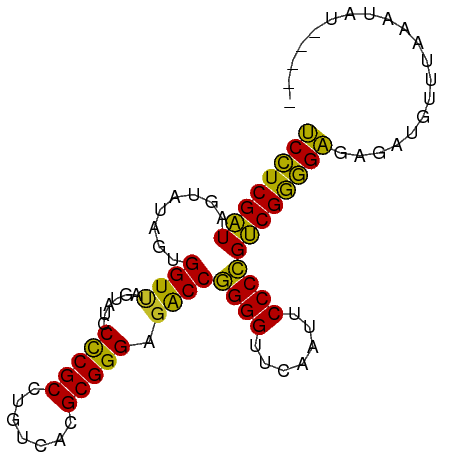
>dm3.chr2L 8541760 87 + 23011544 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUAAAAAUUAC---- ((((((((........((((......(((((.......))))).))))((((.......))))))))))))................---- ( -29.70, z-score = -1.30, R) >droSim1.chr2L 8467835 86 + 22036055 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA-AUGUGAAAACAUU---- ((((((((........((((......(((((.......))))).))))((((.......)))))))))))).(-((((....)))))---- ( -33.20, z-score = -2.24, R) >droSec1.super_3 4028597 87 + 7220098 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA--GAUGUAUAUAUCUU-- ((((((((........((((......(((((.......))))).))))((((.......))))))))))))((--((((....))))))-- ( -32.10, z-score = -1.52, R) >droEre2.scaffold_4929 9136837 86 + 26641161 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGGUGUAUAAUUAU----- .((((..((.((.......)).))(((((.((((.....)))).(((.((((.......))))))))))))))))...........----- ( -30.00, z-score = -0.94, R) >droAna3.scaffold_12916 10186330 84 + 16180835 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGA--UGUUUAAUUAC----- ..............((((((((..(((((.((((.....)))).(((.((((.......))))))))))))..--...))))))))----- ( -29.80, z-score = -1.38, R) >droWil1.scaffold_181071 943347 85 + 1211509 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGU-UAAUUUU----- ((((((((........((((......(((((.......))))).))))((((.......)))))))))))).......-.......----- ( -29.70, z-score = -1.24, R) >droMoj3.scaffold_6500 7168886 81 - 32352404 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAUAUGG-UAC--------- ((((((((........((((......(((((.......))))).))))((((.......)))))))))))).......-...--------- ( -29.70, z-score = -0.80, R) >droGri2.scaffold_15252 9035298 78 - 17193109 GCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACUUGAU------------ (((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))........------------ ( -35.70, z-score = -1.67, R) >droVir3.scaffold_12963 5696917 91 + 20206255 UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAAUGUUCAAAAUAGCUUAU ((((((((........((((......(((((.......))))).))))((((.......)))))))))))).................... ( -29.70, z-score = -1.17, R) >anoGam1.chr3L 4285096 87 + 41284009 UCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGAUUCCCCGUCGGGGAGGGAAAUUAACCUUUU---- ((((((((........((((......(((((.......))))).))))((((.......))))))))))))((........))....---- ( -33.00, z-score = -0.87, R) >consensus UCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAUGUUUAAAUAU_____ ((((((((........((((......(((((.......))))).))))((((.......)))))))))))).................... (-30.53 = -29.56 + -0.97)
| Location | 8,541,760 – 8,541,847 |
|---|---|
| Length | 87 |
| Sequences | 10 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 83.41 |
| Shannon entropy | 0.37397 |
| G+C content | 0.54810 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.96 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
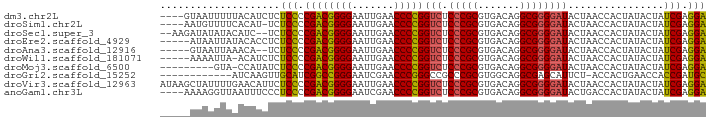
>dm3.chr2L 8541760 87 - 23011544 ----GUAAUUUUUACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA ----................(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -26.80, z-score = -0.87, R) >droSim1.chr2L 8467835 86 - 22036055 ----AAUGUUUUCACAU-UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA ----(((((....))))-).(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -27.20, z-score = -0.86, R) >droSec1.super_3 4028597 87 - 7220098 --AAGAUAUAUACAUC--UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA --.((((......)))--).(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -27.20, z-score = -0.85, R) >droEre2.scaffold_4929 9136837 86 - 26641161 -----AUAAUUAUACACCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA -----...........((((.......(((((.......)))))(((.(((((.......))))))))..................)))). ( -27.60, z-score = -1.27, R) >droAna3.scaffold_12916 10186330 84 - 16180835 -----GUAAUUAAACA--UCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA -----...........--..(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -26.80, z-score = -1.07, R) >droWil1.scaffold_181071 943347 85 - 1211509 -----AAAAUUA-ACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA -----.......-.......(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -26.80, z-score = -1.17, R) >droMoj3.scaffold_6500 7168886 81 + 32352404 ---------GUA-CCAUAUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA ---------...-.......(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -26.80, z-score = -0.68, R) >droGri2.scaffold_15252 9035298 78 + 17193109 ------------AUCAAGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGC ------------........((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))) ( -31.00, z-score = -1.51, R) >droVir3.scaffold_12963 5696917 91 - 20206255 AUAAGCUAUUUUGAACAUUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA ....................(((.((((((((.......)))))(((.(((((.......))))))))................))).))) ( -26.80, z-score = -0.86, R) >anoGam1.chr3L 4285096 87 - 41284009 ----AAAAGGUUAAUUUCCCUCCCCGACGGGGAAUCGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGA ----....(((((...(((((.((((.((((..((((.....))))..)))))).....)).)))))...)))))................ ( -29.60, z-score = -0.42, R) >consensus _____AUAUUUAAACAUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGA ....................(((.((((((((.......)))))(((.(((((.......))))))))................))).))) (-25.85 = -25.43 + -0.42)
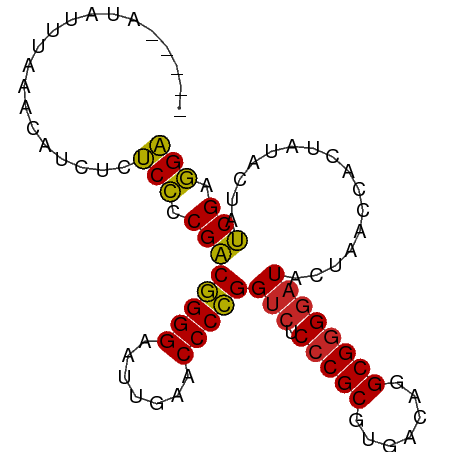
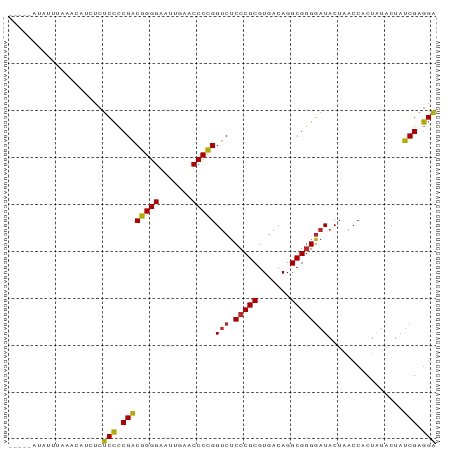
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:16 2011