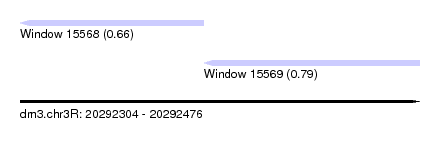
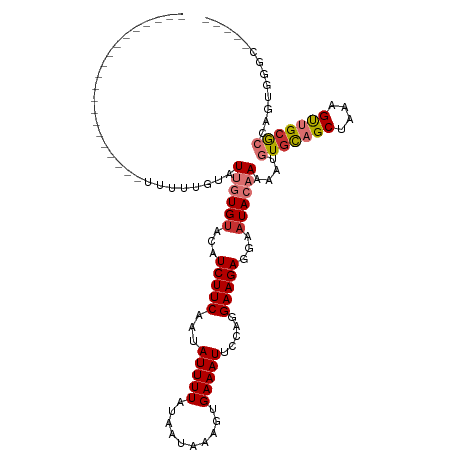
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,292,304 – 20,292,476 |
| Length | 172 |
| Max. P | 0.786081 |

| Location | 20,292,304 – 20,292,383 |
|---|---|
| Length | 79 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 69.98 |
| Shannon entropy | 0.58190 |
| G+C content | 0.49592 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -12.08 |
| Energy contribution | -11.76 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658534 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
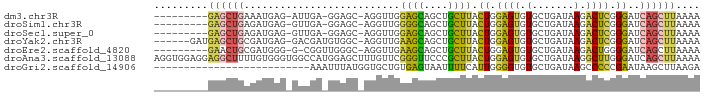
>dm3.chr3R 20292304 79 - 27905053 ---------GAGCUGAAAUGAG-AUUGA-GGAGC-AGGUUGGAGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUCGGGAUCAGCUUAAAA ---------(((((((......-.....-.((((-((.(((...))))))))).((.((((.(.......).)))).)).))))))).... ( -24.20, z-score = -1.87, R) >droSim1.chr3R 20133233 79 - 27517382 ---------GAGCUGAGAUGAG-GUUGA-GGAGC-AGGUUGGGGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUCGGGAUCAGCUUAAAA ---------(((((((......-.....-.((((-((.(((...))))))))).((.((((.(.......).)))).)).))))))).... ( -24.20, z-score = -1.36, R) >droSec1.super_0 20628615 79 - 21120651 ---------GAGCUGAGAUGAG-GUUGA-GGAGC-AGGUUGGAGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUCGGGAUCAGCUUAAAA ---------(((((((......-.....-.((((-((.(((...))))))))).((.((((.(.......).)))).)).))))))).... ( -24.20, z-score = -1.38, R) >droYak2.chr3R 2622199 83 + 28832112 ------GAUGAGCUGCGAUGAG-GACGAUGUGGC-AGGUUGAAGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUCGGGAUCAGCUUAAAA ------.....((..((.((..-..)).))..))-((((((((((....)))..((.((((.(.......).)))).)).))))))).... ( -22.00, z-score = -0.33, R) >droEre2.scaffold_4820 2703000 79 + 10470090 ---------GAACUGCGAUGGG-G-CGGUUGGGC-AGGUUGAAGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUGGGGAUCAGCUUAAAA ---------..(((.((...((-(-((((((..(-.....)...)))))))))..)).)))..((((((..........))))))...... ( -20.70, z-score = -0.07, R) >droAna3.scaffold_13088 333342 91 - 569066 AGGUGGAGGAGGCUUUUGUGGGUGGCCAUGGAGCUUUGUUCGGGUUCCCGCUUACUGGAGUGUGCUGAUAAGGCUUGGGAUCAGCUUAAAA (((((((.((((((.(..(((....)))..))))))).))).(((((((((((....))))).(((.....)))..)))))).)))).... ( -28.90, z-score = -0.36, R) >droGri2.scaffold_14906 314793 65 - 14172833 --------------------------AAAUUUAUGGUGCUGUGAGUAAUUUUCAUUGGGGUGUGCUGAUAAGCCCCCGAAUAAGCUUAAGA --------------------------...........(((.((((.....))))((((((...(((....)))))))))...)))...... ( -14.80, z-score = -0.95, R) >consensus _________GAGCUGAGAUGAG_GUCGA_GGAGC_AGGUUGGAGCAGCUGCUUACUGGAGUGUGCUGAUAAGACUCGGGAUCAGCUUAAAA .........((((((..........................((((....)))).((.((((.(.......).)))).))..)))))).... (-12.08 = -11.76 + -0.32)
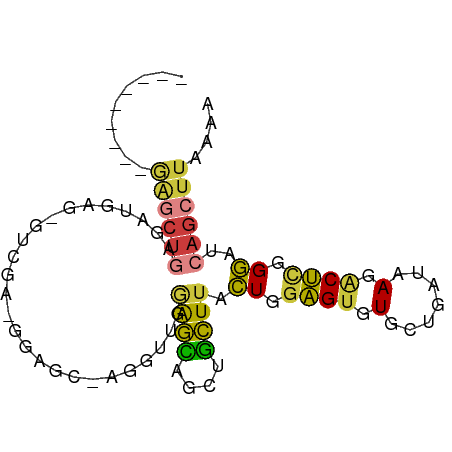
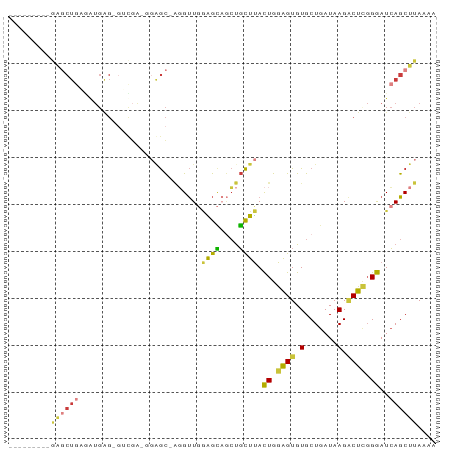
| Location | 20,292,383 – 20,292,476 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Shannon entropy | 0.37416 |
| G+C content | 0.35107 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.34 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.786081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20292383 93 - 27905053 ---------------------UUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCUAAAGUUGCGCCAGUGGGC----- ---------------------.......((((((.(.(((((...(((((((......))))))).....))))).).))))))....(((((((....)))))))........----- ( -22.70, z-score = -2.29, R) >droPer1.super_3 6135752 119 + 7375914 UGUUUUUUUUGUUUUUUGUUUUGUUUCUUCCUGUACAUCUUCAAUAUUUUAUAAUAAAGCGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCAACAGCGGCAACAGCGGCAUCGGG ((((....(((((((((((....((((((((((...........((((....))))...........))))))))))...)))))))...)))).))))((.((....)).))...... ( -24.25, z-score = -0.44, R) >dp4.chr2 12290286 117 + 30794189 --UGUUUUUUUGUUUUUGUUUUGUUUCUUCCUGUACAUCUUCAAUAUUUUAUAAUAAAGCGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGUAGCAACAGCGGCAACAGCGGCAUCGGG --((((....(((((((((....((((((((((...........((((....))))...........))))))))))...)))))))))...))))...((.((....)).))...... ( -26.35, z-score = -1.37, R) >droAna3.scaffold_13088 333433 94 - 569066 --------------------UUGGUGUAUUGUGAACAUCUUCAAUAUUUUAUAAUAAAGCGAAAUUCCAGGAAGAGAAGUACAAAAAUGUGCAGCCAAAGUUGAGCCAGGAGGG----- --------------------(((((...(((((....(((((...((((..(......)..)))).....)))))....))))).......((((....)))).))))).....----- ( -15.80, z-score = 0.03, R) >droEre2.scaffold_4820 2703079 98 + 10470090 ----------------CCUUUUUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCUAAAGUUGCGCCACUGGAC----- ----------------((..((((((((((.(.....(((((...(((((((......))))))).....)))))).)))))))))).(((((((....)))))))....))..----- ( -24.60, z-score = -2.70, R) >droYak2.chr3R 2622282 98 + 28832112 ----------------ACUUAUUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCUGGAAGAGGAAUACAAAAAUGUGCAGCCAAAGUUGCGCCAGCGGAC----- ----------------......(((((.((((((.(.(((((...(((((((......))))))).....))))).).))))))....(((((((....)))))))..))))).----- ( -23.80, z-score = -2.24, R) >droSec1.super_0 20628694 93 - 21120651 ---------------------UUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCUAAAGUUGCGCCAGUGGGC----- ---------------------.......((((((.(.(((((...(((((((......))))))).....))))).).))))))....(((((((....)))))))........----- ( -22.70, z-score = -2.29, R) >droSim1.chr3R 20133312 93 - 27517382 ---------------------UUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCUAAAGUUGCGCCAGUGGGC----- ---------------------.......((((((.(.(((((...(((((((......))))))).....))))).).))))))....(((((((....)))))))........----- ( -22.70, z-score = -2.29, R) >consensus ____________________UUUUUGUAUUGUGUACAUCUUCAAUAUUUUAUAAUAAAGUGAAAUUCCAGGAAGAGGAAUACAAAAAUGUGCAGCUAAAGUUGCGCCAGUGGGC_____ ............................((((((...(((((...(((((..........))))).....)))))...))))))....(((((((....)))))))............. (-14.57 = -15.34 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:47 2011